-
摘要
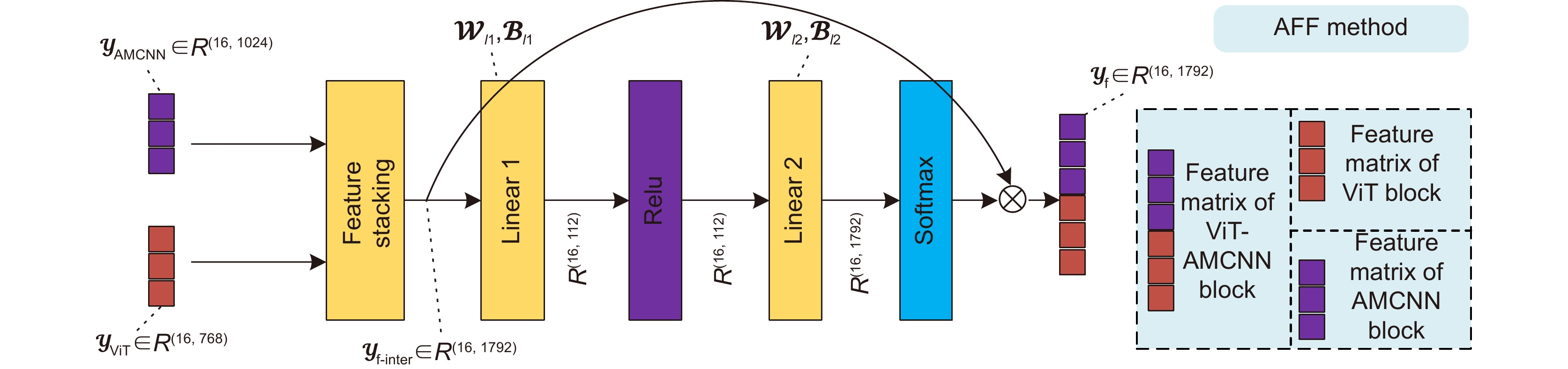
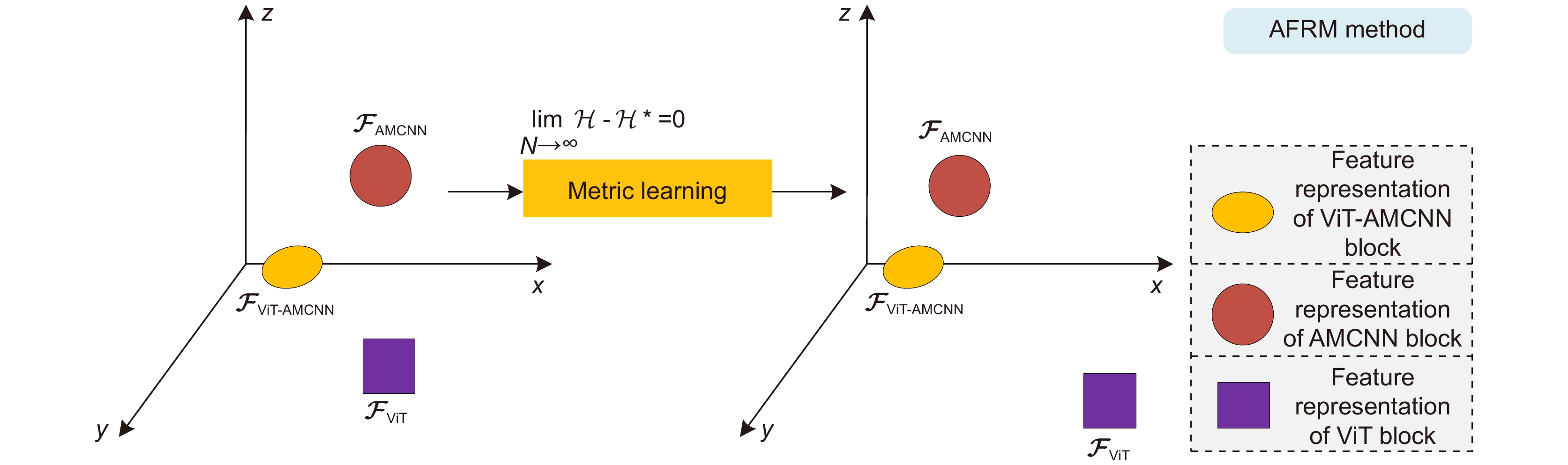
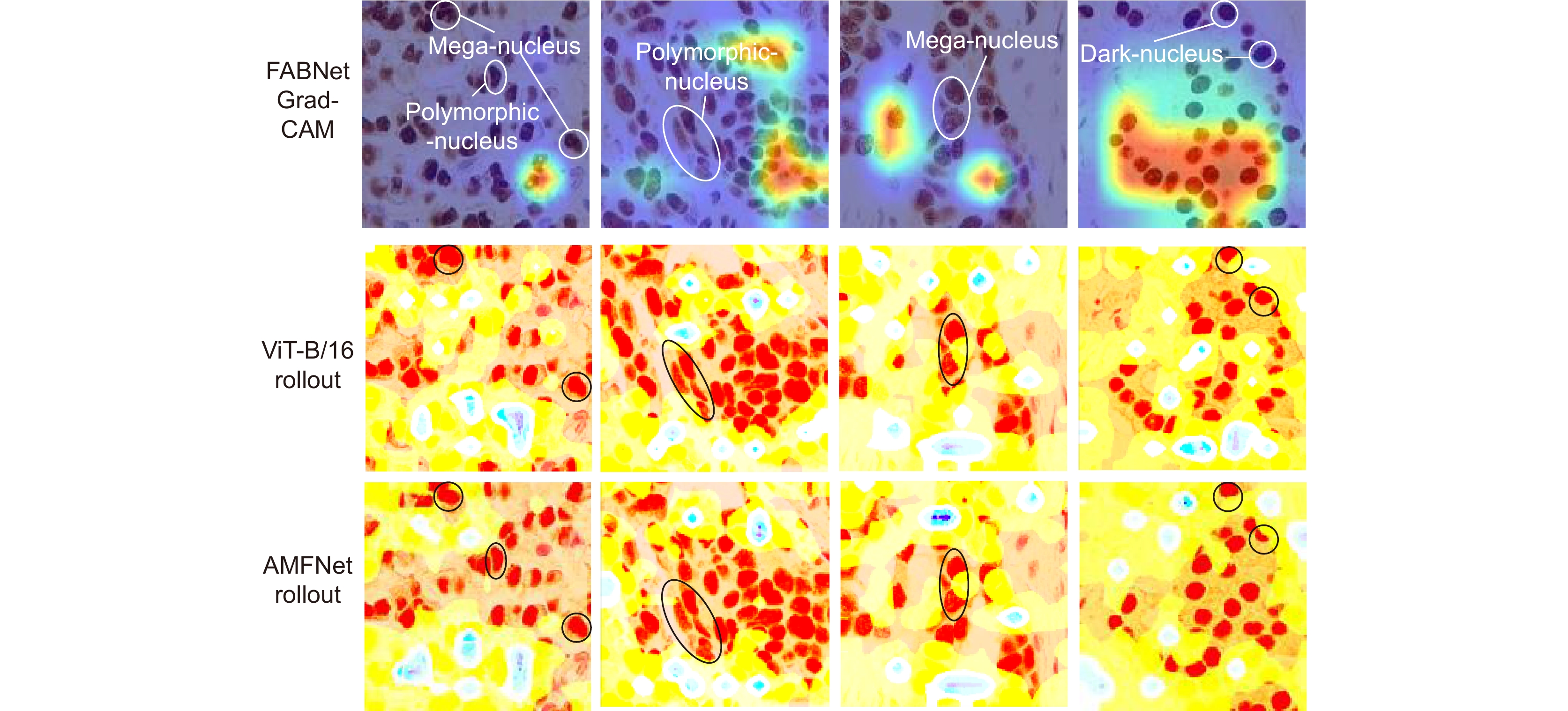
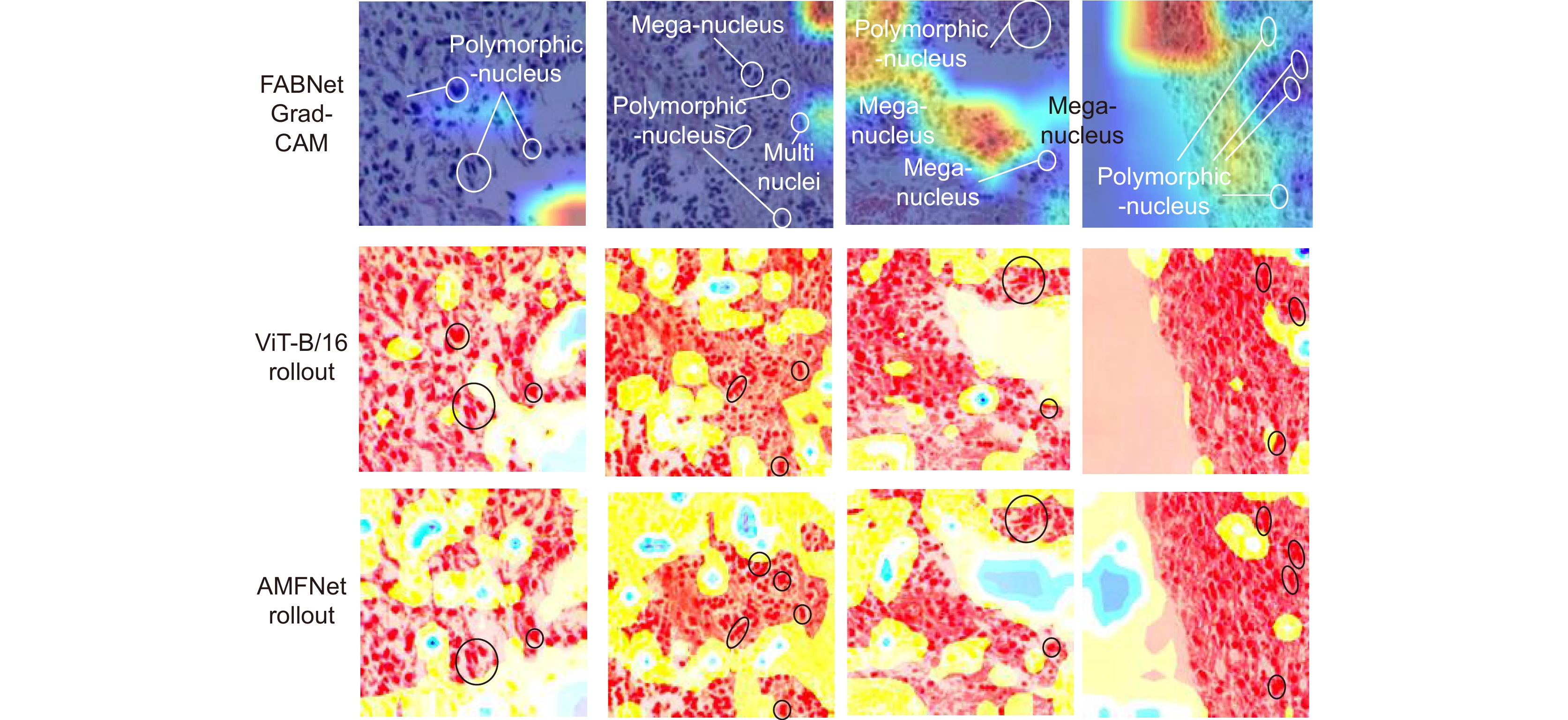
基于显微成像技术的肿瘤分级对于乳腺癌诊断和预后有着重要的意义, 且诊断结果需具备高精度和可解释性。目前,集成Attention的CNN模块深度网络归纳偏差能力较强,但可解释性较差;而基于ViT块的深度网络其可解释性较好,但归纳偏差能力较弱。本文通过融合ViT块和集成Attention的CNN块,提出了一种端到端的自适应模型融合的深度网络。由于现有模型融合方法存在负融合现象,无法保证ViT块和集成Attention的CNN块同时具有良好的特征表示能力;另外,两种特征表示之间相似度高且冗余信息多,导致模型融合能力较差。为此,本文提出一种包含多目标优化、自适应特征表示度量和自适应特征融合的自适应模型融合方法,有效地提高了模型的融合能力。实验表明本文模型的准确率达到95.14%, 相比ViT-B/16提升了9.73%,比FABNet提升了7.6%;模型的可视化图更加关注细胞核异型的区域(例如巨型核、多形核、多核和深色核),与病理专家所关注的区域更加吻合。整体而言,本文所提出的模型在精度和可解释性上均优于当前最先进的(state of the art)模型。
Abstract
Tumor grading based on microscopic imaging is critical for the diagnosis and prognosis of breast cancer, which demands excellent accuracy and interpretability. Deep networks with CNN blocks combined with attention currently offer better induction bias capabilities but low interpretability. In comparison, the deep network based on ViT blocks has stronger interpretability but less induction bias capabilities. To that end, we present an end-to-end adaptive model fusion for deep networks that combine ViT and CNN blocks with integrated attention. However, the existing model fusion methods suffer from negative fusion. Because there is no guarantee that both the ViT blocks and the CNN blocks with integrated attention have acceptable feature representation capabilities, and secondly, the great similarity between the two feature representations results in a lot of redundant information, resulting in a poor model fusion capability. For that purpose, the adaptive model fusion approach suggested in this study consists of multi-objective optimization, an adaptive feature representation metric, and adaptive feature fusion, thereby significantly boosting the model's fusion capabilities. The accuracy of this model is 95.14%, which is 9.73% better than that of ViT-B/16, and 7.6% better than that of FABNet; secondly, the visualization map of our model is more focused on the regions of nuclear heterogeneity (e.g., mega nuclei, polymorphic nuclei, multi-nuclei, and dark nuclei), which is more consistent with the regions of interest to pathologists. Overall, the proposed model outperforms other state-of-the-art models in terms of accuracy and interpretability.
-
Key words:
- microscopic imaging /
- interpretability /
- deep learning /
- adaptive fusion /
- breast cancer /
- tumor grading
-
Overview
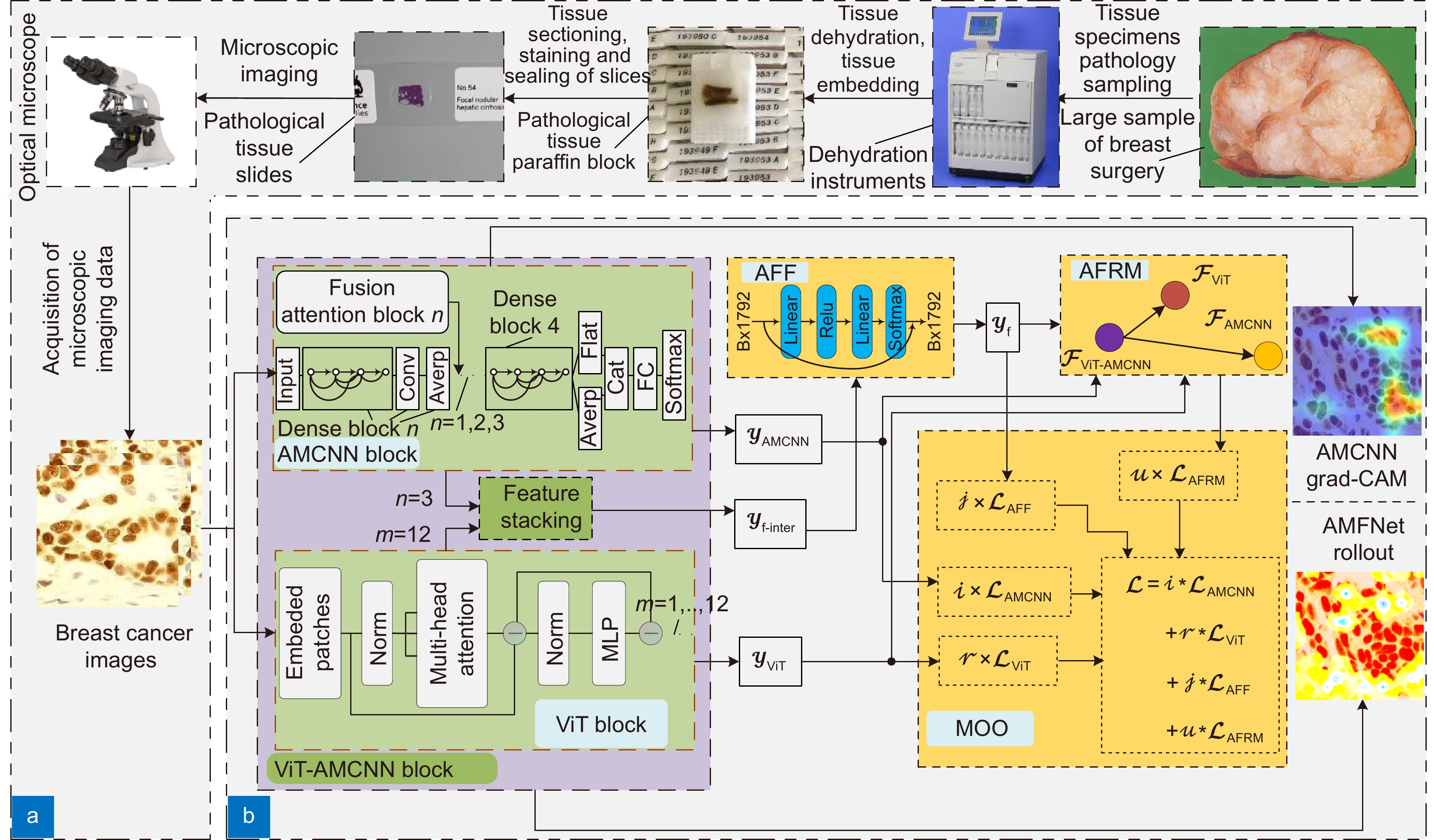
Overview: Breast cancer is the most common cancer among women. Tumor grading based on microscopic imaging is important for the diagnosis and prognosis of breast cancer, and the results need to be highly accurate and interpretable. Breast cancer tumor grading relies on pathologists to assess the morphological status of tissues and cells in the microscopic images of tissue sections, such as tissue differentiation, nuclear isotypes, and mitotic counts. A strong statistical correlation between the hematoxylin-Eosin (HE) stained microscopic imaging samples and the progesterone Receptor (ER) immunohistochemically (IHC) stained microscopic imaging samples has been documented, i.e., the ER status is strongly correlated with the tumor tissue grading. Therefore, it is a meaningful task to use deep learning models to research the breast tumor grading in ER IHC pathology images exploratively. At present, the CNN module integrating attention has a strong ability of induction bias but poor interpretability, while the Vision Transformer (ViT) block-based deep network has better interpretability but weaker ability of induction bias. In this paper, we propose an end-to-end deep network with adaptive model fusion by fusing ViT blocks and CNN blocks with integrated attention. Due to the negative fusion phenomenon of the existing model fusion methods, while it is impossible to guarantee that ViT blocks and CNN blocks with integrated attention have good feature representation capability at the same time; in addition, the high similarity and redundant information between the two feature representations lead to a poor model fusion capability. To this end, this paper proposes an adaptive model fusion method that includes multi-objective optimization, adaptive feature representation metric, and adaptive feature fusion, which effectively improves the fusion ability of the model. The accuracy of the model is 95.14%, which is 9.73% better than that of ViT-B/16 and 7.6% better than that of FABNet. The visualization of the model focuses more on the regions of nuclear heterogeneity (e.g., giant nuclei, polymorphic nuclei, multinuclei and dark nuclei), which is more consistent with the regions of interest to pathologists. Overall, the proposed model outperforms the current state-of-the-art model in terms of accuracy and interpretability.
-
-
表 1 乳腺癌ER IHC数据集数量分布表
Table 1. Distribution of the number of ER IHC datasets for breast cancer
Datasets Parameter Grade I Grade II Grade III Image size Total Training set 268 355 360 224×224 983 Validation set 90 118 120 224×224 328 Testing set 90 119 120 224×224 329 Total 448 592 600 224×224 1640 表 2 分类混淆矩阵
Table 2. Classification confusion matrix
Reality Forecast result Positive Negative Positive True positive (TP) False negative (FN) Negative False positive (FP) True negative (TN) 表 3 在ER IHC染色的乳腺癌病理组织显微镜成像中对AMF方法进行消融 (✗表示无,✓表示有)
Table 3. Ablation of AMF method in ER IHC pathological microimaging of breast cancer
Model AMF method Average acc/% P R F1 AUC MOO AFRM AFF AMFNet (ViT-AMCNN blocks) ✗ ✗ ✓ 69.00 0.6934 0.6900 0.6903 0.7643 ✓ ✗ ✗ 92.40 0.9240 0.9240 0.9240 0.9423 ✓ ✓ ✗ 93.92 0.9403 0.9392 0.9394 0.9532 ✓ ✓ ✓ 95.14 0.9520 0.9514 0.9513 0.9617 表 4 乳腺癌ER IHC染色的病理组织显微成像的肿瘤分级准确率
Table 4. Tumor grading accuracy of breast cancer ER IHC histopathology microscopic imaging
Model Grade I acc/% Grade II acc/% Grade III acc/% Average acc/% P R F1 AUC Inception V3[36] 74.71 78.19 78.01 77.20 0.7721 0.7720 0.7717 0.8290 Xception V3[37] 71.82 68.40 76.42 72.34 0.7226 0.7234 0.7226 0.7925 ResNet50[38] 72.96 73.77 76.08 74.47 0.7524 0.7447 0.7439 0.8085 DenseNet121[39] 82.80 77.53 81.63 80.55 0.8059 0.8055 0.8047 0.8541 DenseNet121+Nonlocal[40] 85.88 84.39 81.97 83.89 0.8396 0.8389 0.8391 0.8791 DenseNet121+SENet[41] 79.55 83.27 84.39 82.67 0.8272 0.8267 0.8266 0.8700 DenseNet121+CBAM[42] 82.76 81.20 84.80 82.98 0.8307 0.8298 0.8294 0.8723 DenseNet121+HIENet[43] 86.21 85.59 85.48 85.71 0.8585 0.8571 0.8572 0.8928 FABNet[15] 86.05 91.29 84.90 87.54 0.8765 0.8754 0.8752 0.9036 ViT-S/16[27] 54.02 55.87 69.20 60.18 60.37 60.18 0.6023 0.6970 ViT-B/16[27] 85.87 86.32 84.17 85.41 0.8546 0.8541 0.8541 0.8913 ViT-B/32[27] 68.60 70.00 73.98 71.12 0.7114 0.7112 0.7107 0.7799 ViT-L/16[27] 78.98 79.17 81.23 79.94 0.8113 0.7994 0.7987 0.8430 ViT-L/32[27] 50.35 61.21 59.83 58.36 0.6018 0.5836 0.5774 0.6770 AMFNet (ours) 92.66 97.02 95.12 95.14$ \uparrow $7.6 0.9520 0.9514 0.9513 0.9617 表 5 脑癌组织病理图像数据集数量分布表
Table 5. Distribution of the number of brain cancer histopathology image datasets
Datasets Grade I Grade II Grade III Grade IV Total Training set 276 492 873 891 2532 Validation set 91 164 290 296 841 Testing set 91 164 290 296 841 Total 458 820 1453 1483 4214 表 6 脑癌组织病理图像的肿瘤分级准确率的对比表
Table 6. Comparison table of tumor grading accuracy of histopathological images of brain cancer
Model Metrics Grade I acc/% Grade II acc/% Grade III acc/% Grade IV acc/% Average acc/% P R F1 AUC Inception V3[36] 84.16 68.11 80.62 78.05 77.76 0.7779 0.7776 0.7766 0.8575 Xception V3[37] 84.95 72.34 78.31 80.80 78.83 0.7976 0.7883 0.7874 0.8588 ResNet50[38] 82.00 65.62 66.43 71.12 69.80 0.6963 0.6980 0.6961 0.8091 DenseNet121[39] 87.05 74.75 74.86 81.75 78.95 0.7978 0.7895 0.7858 0.8625 DenseNet121+Nonlocal[40] 91.84 81.90 86.70 89.39 87.40 0.8763 0.8740 0.8727 0.9195 DenseNet121+SENet[41] 96.22 84.97 88.05 90.25 89.18 0.8925 0.8918 0.8911 0.9279 DenseNet121+CBAM[42] 92.71 82.00 89.04 86.77 87.40 0.8750 0.8740 0.8726 0.9162 DenseNet121+HIENet[43] 95.70 85.99 87.81 87.50 88.23 0.8851 0.8823 0.8820 0.9244 FABNet[15] 93.68 87.70 90.97 90.82 90.61 0.9072 0.9061 0.9057 0.9391 ViT-S/16[27] 65.98 46.98 65.97 69.73 64.21 0.6375 0.6421 0.6359 0.7489 ViT-B/16[27] 83.90 80.00 87.83 86.83 85.61 0.8618 0.8561 0.8553 0.9028 ViT-B/32[27] 81.48 62.59 78.93 77.20 75.74 0.7550 0.7574 0.7541 0.8319 ViT-L/16[27] 70.79 54.49 72.05 73.56 69.32 0.6897 0.6932 0.6902 0.7808 ViT-L/32[27] 75.49 58.02 73.76 75.08 71.70 0.7152 0.7170 0.7134 0.8084 AMFNet (ours) 98.32 93.38 94.30 93.90 94.41$ \uparrow $3.8 0.9451 0.9441 0.9442 0.9611 -
参考文献
[1] Parkin D M, Bray F, Ferlay J, et al. Estimating the world cancer burden: globocan 2000[J]. Int J Cancer, 2001, 94(2): 153−156. doi: 10.1002/ijc.1440
[2] Robertson S, Azizpour H, Smith K, et al. Digital image analysis in breast pathology—from image processing techniques to artificial intelligence[J]. Transl Res, 2018, 194: 19−35. doi: 10.1016/j.trsl.2017.10.010
[3] Desai S B, Moonim M T, Gill A K, et al. Hormone receptor status of breast cancer in India: a study of 798 tumours[J]. Breast, 2000, 9(5): 267−270. doi: 10.1054/brst.2000.0134
[4] Zafrani B, Aubriot M H, Mouret E, et al. High sensitivity and specificity of immunohistochemistry for the detection of hormone receptors in breast carcinoma: comparison with biochemical determination in a prospective study of 793 cases[J]. Histopathology, 2000, 37(6): 536−545. doi: 10.1046/j.1365-2559.2000.01006.x
[5] Fuqua S A W, Schiff R, Parra I, et al. Estrogen receptor β protein in human breast cancer: correlation with clinical tumor parameters[J]. Cancer Res, 2003, 63(10): 2434−2439.
[6] Vagunda V, Šmardová J, Vagundová M, et al. Correlations of breast carcinoma biomarkers and p53 tested by FASAY and immunohistochemistry[J]. Pathol Res Pract, 2003, 199(12): 795−801. doi: 10.1078/0344-0338-00498
[7] Baqai T, Shousha S. Oestrogen receptor negativity as a marker for high-grade ductal carcinoma in situ of the breast[J]. Histopathology, 2003, 42(5): 440−447. doi: 10.1046/j.1365-2559.2003.01612.x
[8] Sofi G N, Sofi J N, Nadeem R, et al. Estrogen receptor and progesterone receptor status in breast cancer in relation to age, histological grade, size of lesion and lymph node involvement[J]. Asian Pac J Cancer Prev, 2012, 13(10): 5047−5052. doi: 10.7314/apjcp.2012.13.10.5047
[9] Azizun-Nisa, Bhurgri Y, Raza F, et al. Comparison of ER, PR and HER-2/neu (C-erb B 2) reactivity pattern with histologic grade, tumor size and lymph node status in breast cancer[J]. Asian Pac J Cancer Prev, 2008, 9(4): 553−556.
[10] Gurcan M N, Boucheron L E, Can A, et al. Histopathological image analysis: a review[J]. IEEE Rev Biomed Eng, 2009, 2: 147−171. doi: 10.1109/RBME.2009.2034865
[11] Yang H, Kim J Y, Kim H, et al. Guided soft attention network for classification of breast cancer histopathology images[J]. IEEE Trans Med Imaging, 2020, 39(5): 1306−1315. doi: 10.1109/TMI.2019.2948026
[12] BenTaieb A, Hamarneh G. Predicting cancer with a recurrent visual attention model for histopathology images[C]//21st International Conference on Medical Image Computing and Computer Assisted Intervention, 2018: 129–137. https://doi.org/10.1007/978-3-030-00934-2_15.
[13] Tomita N, Abdollahi B, Wei J, et al. Attention-based deep neural networks for detection of cancerous and precancerous esophagus tissue on histopathological slides[J]. JAMA Netw Open, 2019, 2(11): e1914645. doi: 10.1001/jamanetworkopen.2019.14645
[14] Yao J W, Zhu X L, Jonnagaddala J, et al. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks[J]. Med Image Anal, 2020, 65: 101789. doi: 10.1016/j.media.2020.101789
[15] Huang P, Tan X H, Zhou X L, et al. FABNet: Fusion attention block and transfer learning for laryngeal cancer tumor grading in p63 IHC histopathology images[J]. IEEE J Biomed Health Inform, 2022, 26(4): 1696−1707. doi: 10.1109/JBHI.2021.3108999
[16] Zhou X L, Tang C W, Huang P, et al. LPCANet: classification of laryngeal cancer histopathological images using a CNN with position attention and channel attention mechanisms[J]. Interdiscip Sci, 2021, 13(4): 666−682. doi: 10.1007/s12539-021-00452-5
[17] 刘敬璇, 樊金宇, 汪权, 等. SS-OCTA对黑色素瘤皮肤结构和血管的成像实验[J]. 光电工程, 2020, 47(2): 190239. doi: 10.12086/oee.2020.190239
Liu J X, Fan J Y, Wang Q, et al. Imaging of skin structure and vessels in melanoma by swept source optical coherence tomography angiography[J]. Opto-Electron Eng, 2020, 47(2): 190239. doi: 10.12086/oee.2020.190239
[18] Abnar S, Zuidema W. Quantifying attention flow in transformers[C]//Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, 2020: 4190–4197. https://doi.org/10.18653/v1/2020.acl-main.385.
[19] Chefer H, Gur S, Wolf L. Transformer interpretability beyond attention visualization[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2021: 782–791. https://doi.org/10.1109/CVPR46437.2021.00084.
[20] 梁礼明, 周珑颂, 陈鑫, 等. 鬼影卷积自适应视网膜血管分割算法[J]. 光电工程, 2021, 48(10): 210291. doi: 10.12086/oee.2021.210291
Liang L M, Zhou L S, Chen X, et al. Ghost convolution adaptive retinal vessel segmentation algorithm[J]. Opto-Electron Eng, 2021, 48(10): 210291. doi: 10.12086/oee.2021.210291
[21] 刘侠, 甘权, 刘晓, 等. 基于超像素的联合能量主动轮廓CT图像分割方法[J]. 光电工程, 2020, 47(1): 190104. doi: 10.12086/oee.2020.190104
Liu X, Gan Q, Liu X, et al. Joint energy active contour CT image segmentation method based on super-pixel[J]. Opto-Electron Eng, 2020, 47(1): 190104. doi: 10.12086/oee.2020.190104
[22] Gao Z Y, Hong B Y, Zhang X L, et al. Instance-based vision transformer for subtyping of papillary renal cell carcinoma in histopathological image[C]//24th International Conference on Medical Image Computing and Computer Assisted Intervention, 2021: 299–308. https://doi.org/10.1007/978-3-030-87237-3_29.
[23] Wang X Y, Yang S, Zhang J, et al. TransPath: Transformer-based self-supervised learning for histopathological image classification[C]//24th International Conference on Medical Image Computing and Computer Assisted Intervention, 2021: 186–195. https://doi.org/10.1007/978-3-030-87237-3_18.
[24] Li H, Yang F, Zhao Y, et al. DT-MIL: deformable transformer for multi-instance learning on histopathological image[C]//24th International Conference on Medical Image Computing and Computer Assisted Intervention, 2021: 206–216. https://doi.org/10.1007/978-3-030-87237-3_20.
[25] Chen H Y, Li C, Wang G, et al. GasHis-transformer: a multi-scale visual transformer approach for gastric histopathological image detection[J]. Pattern Recognit, 2022, 130: 108827. doi: 10.1016/j.patcog.2022.108827
[26] Zou Y, Chen S N, Sun Q L, et al. DCET-Net: dual-stream convolution expanded transformer for breast cancer histopathological image classification[C]//2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2021: 1235–1240. https://doi.org/10.1109/BIBM52615.2021.9669903.
[27] Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: transformers for image recognition at scale[C]//9th International Conference on Learning Representations, 2021.
[28] Breiman L. Bagging predictors[J]. Mach Learn, 1996, 24(2): 123−140. doi: 10.1023/A:1018054314350
[29] Schapire R E. The boosting approach to machine learning: an overview[M]//Denison D D, Hansen M H, Holmes C C, et al. Nonlinear Estimation and Classification. New York: Springer, 2003: 149–171. https://doi.org/10.1007/978-0-387-21579-2_9.
[30] Ting K M, Witten I H. Stacking bagged and Dagged models[C]//Proceedings of the Fourteenth International Conference on Machine Learning, 1997: 367–375. https://doi.org/10.5555/645526.657147.
[31] Ke W C, Fan F, Liao P X, et al. Biological gender estimation from panoramic dental X-ray images based on multiple feature fusion model[J]. Sens Imaging, 2020, 21(1): 54. doi: 10.1007/s11220-020-00320-4
[32] 周志华. 机器学习[M]. 北京: 清华大学出版社, 2016: 171–196.
Zhou Z H. Machine Learning[M]. Beijing: Tsinghua University Press, 2016: 171–196.
[33] Selvaraju R R, Cogswell M, Das A, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization[C]//Proceedings of the 2017 IEEE International Conference on Computer Vision, 2017: 618–626.
[34] Selvaraju R R, Cogswell M, Das A, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization[C]//Proceedings of the IEEE International Conference on Computer Vision, 2017: 618–626. https://doi.org/10.1109/ICCV.2017.74.
[35] Kostopoulos S, Glotsos D, Cavouras D, et al. Computer-based association of the texture of expressed estrogen receptor nuclei with histologic grade using immunohistochemically-stained breast carcinomas[J]. Anal Quant Cytol Histol, 2009, 31(4): 187−196.
[36] Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the inception architecture for computer vision[C]//2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016: 2818–2826. https://doi.org/10.1109/CVPR.2016.308.
[37] Chollet F. Xception: deep learning with depthwise separable convolutions[C]//30th IEEE Conference on Computer Vision and Pattern Recognition, 2017: 1800–1807. https://doi.org/10.1109/CVPR.2017.195.
[38] He K M, Zhang X Y, Ren S Q, et al. Deep residual learning for image recognition[C]//2016 IEEE Conference on Computer Vision and Pattern Recognition, 2016: 770–778. https://doi.org/10.1109/CVPR.2016.90.
[39] Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017: 2261–2269. https://doi.org/10.1109/CVPR.2017.243.
[40] Wang X L, Girshick R, Gupta A, et al. Non-local neural networks[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018: 7794–7803. https://doi.org/10.1109/CVPR.2018.00813.
[41] Hu J, Shen L, Albanie S, et al. Squeeze-and-excitation networks[J]. IEEE Trans Pattern Anal Mach Intell, 2020, 42(8): 2011−2023. doi: 10.1109/TPAMI.2019.2913372
[42] Woo S, Park J, Lee J Y, et al. CBAM: convolutional block attention module[C]//Proceedings of the 15th European Conference on Computer Vision, 2018: 3–19. https://doi.org/10.1007/978-3-030-01234-2_1.
[43] Sun H, Zeng X X, Xu T, et al. Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms[J]. IEEE J Biomed Health Inform, 2020, 24(6): 1664−1676. doi: 10.1109/JBHI.2019.2944977
[44] Glotsos D, Kalatzis I, Spyridonos P, Kostopoulos S, Daskalakis A, Athanasiadis E, Ravazoula P, Nikiforidis G, and Cavouras D. Improving accuracy in astrocytomas grading by integrating a robust least squares mapping driven support vector machine classifier into a two level grade classification scheme[J]. Comput Methods Programs Biomed, 2008, 90(3): 251−261. doi: 10.1016/j.cmpb.2008.01.006
-
访问统计

 E-mail Alert
E-mail Alert RSS
RSS
 下载:
下载: