Intravascular ultrasound image segmentation combining polar coordinate modeling and a neural network
-
摘要
针对现有血管内超声(IVUS)图像分割网络不能保证分割结果之间的拓扑关系符合医学先验知识,影响后续临床参数计算的问题,提出了一种基于极坐标建模和密集距离回归网络的IVUS图像分割方法。首先通过极坐标建模将含有先验知识的二维掩膜编码为一维距离向量;然后构建一个结合残差网络和语义嵌入模块的密集距离回归网络,用于学习IVUS图像和一维距离向量之间的映射关系。同时提出联合损失函数约束网络的学习方向。预测结果最终通过样条曲线拟合被重建为二维掩模。实验结果表明,所提方法在血管、管腔和斑块区域的分割结果拓扑关系100%符合先验知识,Jaccard测量值分别达到0.89、0.87和0.74。该算法适用于一般的IVUS图像分割,分割结果中血管结构定位准确,拓扑关系正确,可提供可靠的临床参数。
Abstract
Aiming at the problem that existing intravascular ultrasound (IVUS) image segmentation networks cannot guarantee that the topological relationships between segmentation results conform to medical prior knowledge, which has a negative impact on clinical parameter calculation, an IVUS image segmentation method based on polar coordinate modeling and dense-distance regression network is proposed. This method converts two-dimensional (2D) masks to one-dimensional (1D) distance vectors to preserve the topology of the vessel structures through polar coordinate modeling with prior knowledge. Then a dense-distance regression network consisting of a residual network and semantic embedding branch is constructed for learning the mapping relationships between IVUS images and 1D distance vectors. A joint loss function is proposed to constrain the network learning direction. The prediction results are finally reconstructed as 2D masks by spline curve fitting. The experimental results show that the proposed method achieves 100% topology preservation in the media, lumen, and plaque regions, and achieves Jaccard measure (JM) of 0.89, 0.87, and 0.74, respectively. The algorithm is suitable for general IVUS image segmentation, with high accuracy, and can provide reliable clinical parameters.
-
Overview
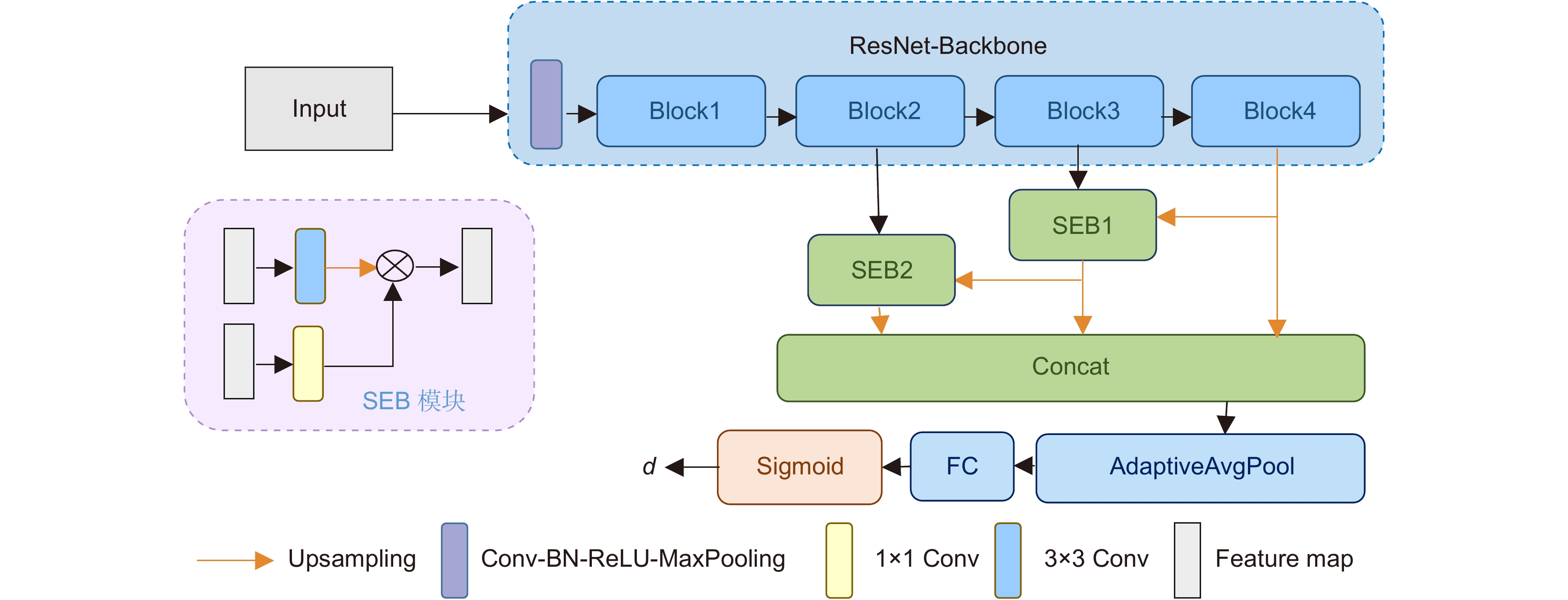
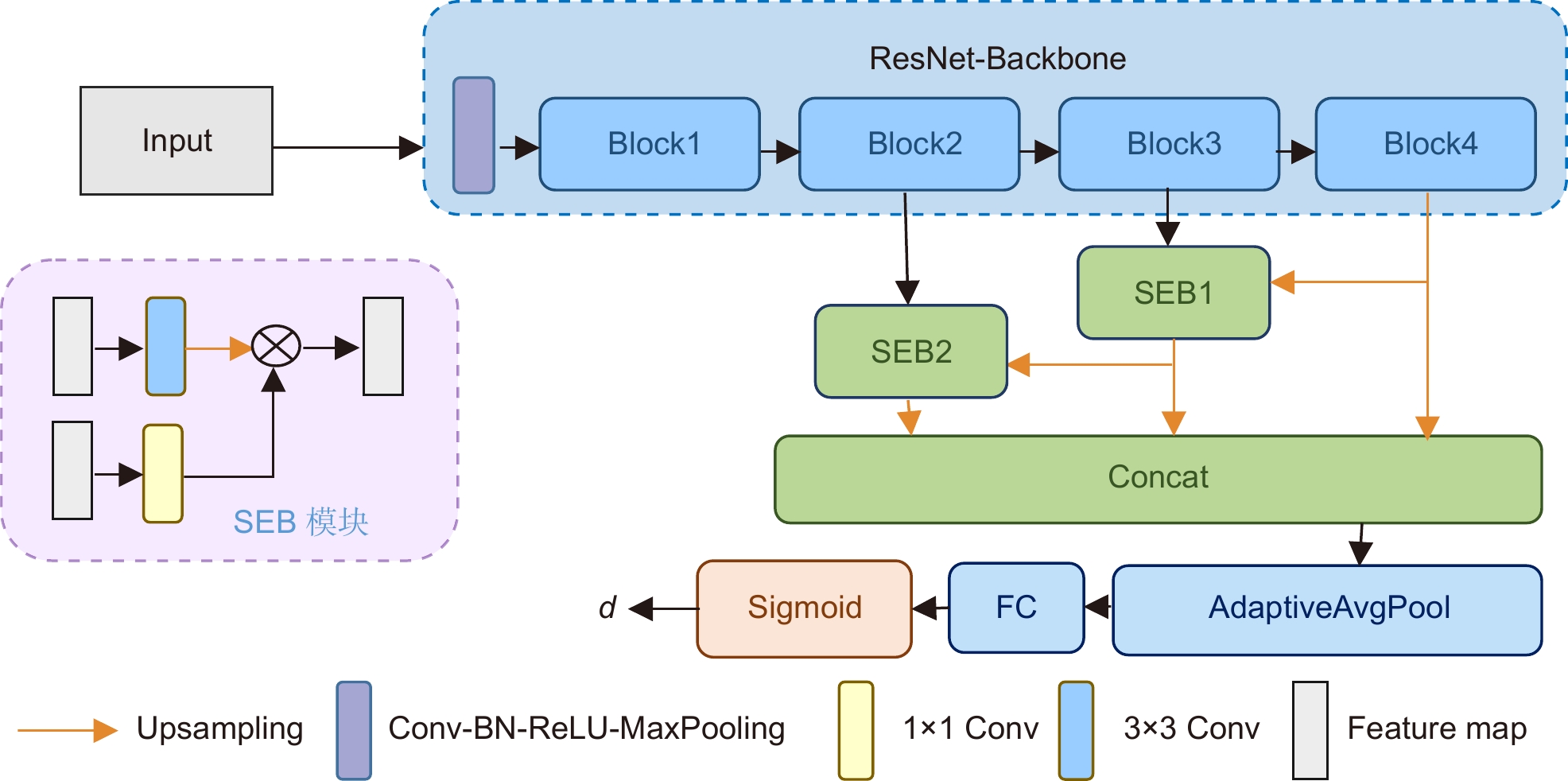
Overview: Intravascular ultrasound (IVUS) is an important imaging modality for diagnosing cardiovascular diseases. The annotation of major anatomical structures of blood vessels in IVUS images can provide necessary clinical parameters for lesion severity assessment, which is a necessary step for physicians' diagnosis. However, manual annotation is laborious and inefficient. With the development of deep learning, convolutional neural networks perform well in this task and are able to achieve automatic and accurate segmentation and recognition of the main anatomical structures of blood vessels. Existing IVUS image segmentation networks are mostly based on pixel-by-pixel prediction, which lacks overall constraints on the main structures of blood vessels and cannot guarantee that the topological relationships between main vessel structures conform to medical prior knowledge, which has a negative impact on the calculation of clinical parameters. To solve this problem, this paper proposes an IVUS image segmentation method based on polar coordinate modeling and a dense-distance regression network. First, a prior knowledge-based polar coordinate modeling is designed for encoding the two-dimensional mask of the main structure of blood vessels containing prior knowledge into a one-dimensional distance vector to avoid the topological relationship of the blood vessel structure from generating random changes in the network prediction. A dense-distance regression network consisting of a residual network and a semantic embedding branching module is then constructed for learning the mapping relationship between IVUS images and 1D distance vectors. To effectively constrain the learning direction of the network, a joint loss function is proposed. This loss function takes into account the actual spatial relationship between one-dimensional distance vectors and has a stronger supervisory capability. The network prediction results are finally reconstructed as a two-dimensional mask by spline curve fitting. The proposed method is validated on a 20 MHz IVUS image dataset. The experimental results show that the proposed method achieves 100% topology preservation in the media, lumen, and plaque regions and achieves the Jaccard measure (JM) of 0.89, 0.87, and 0.74, respectively. The advantage of the algorithm in this paper is that it can provide a high accuracy and topologically correct segmentation results of the vessel structures, which is suitable for general IVUS image segmentation. The clinical parameters provided are reliable and can be used as an important reference basis for physicians' diagnosis, reducing physicians' workload and improving diagnostic efficiency, which has a promising future in clinical applications.
-
-
图 2 建模示意图。 (a) IVUS 原图;(b) 建模结果示意图。内膜边界和中-外膜边界分别用红色和绿色曲线标记,管腔区域和斑块区域建模结果分别使用红色和绿色线段标记
Figure 2. Modeling schematics. (a) Original image of IVUS; (b) Schematic diagram of modeling result. The intima contour and media contour are marked with red and green curves, respectively. The modeling results of the lumen area and plaque area are marked with red and green line segments, respectively
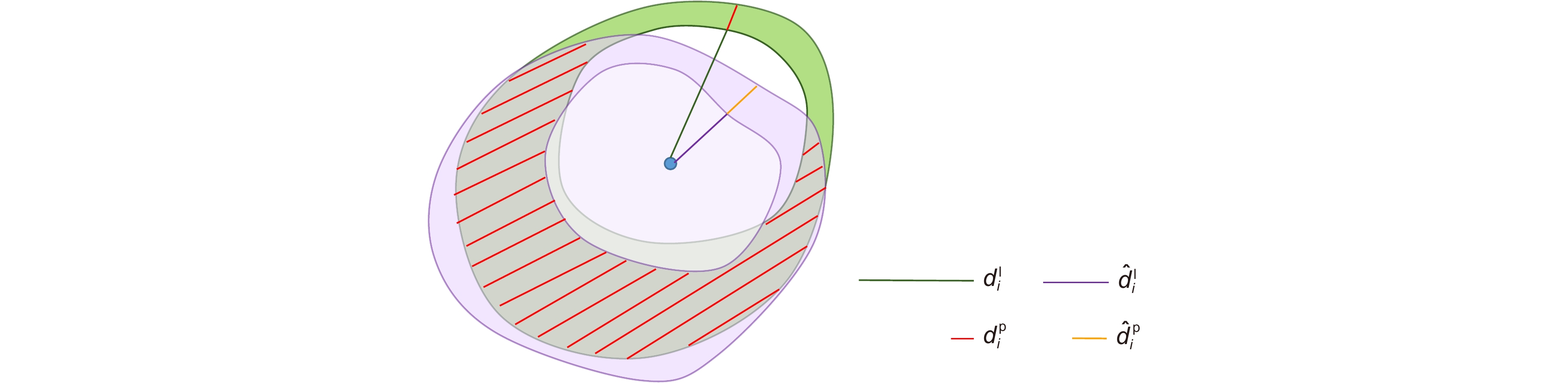
图 4 真值与预测值斑块区域交并情况示意图。注:为便于观察,将真值射线与预测值射线错开一定角度表示,实际二者在同一射线上
Figure 4. Schematic diagram of the intersection of the true value and the predicted value patch area. Note: For the convenience of observation, the true value ray and the predicted value ray are staggered by a certain angle, and the two are actually on the same ray
表 1 IVUS数据集信息
Table 1. Information of the IVUS dataset
患者标号 1 2 3 4 图像数量 218 39 318 168 表 2 不同深度的骨架网络与不同数量SEB模块组合实验结果
Table 2. The performance of the proposed method under different depths of backbone and different numbers of SEB modules
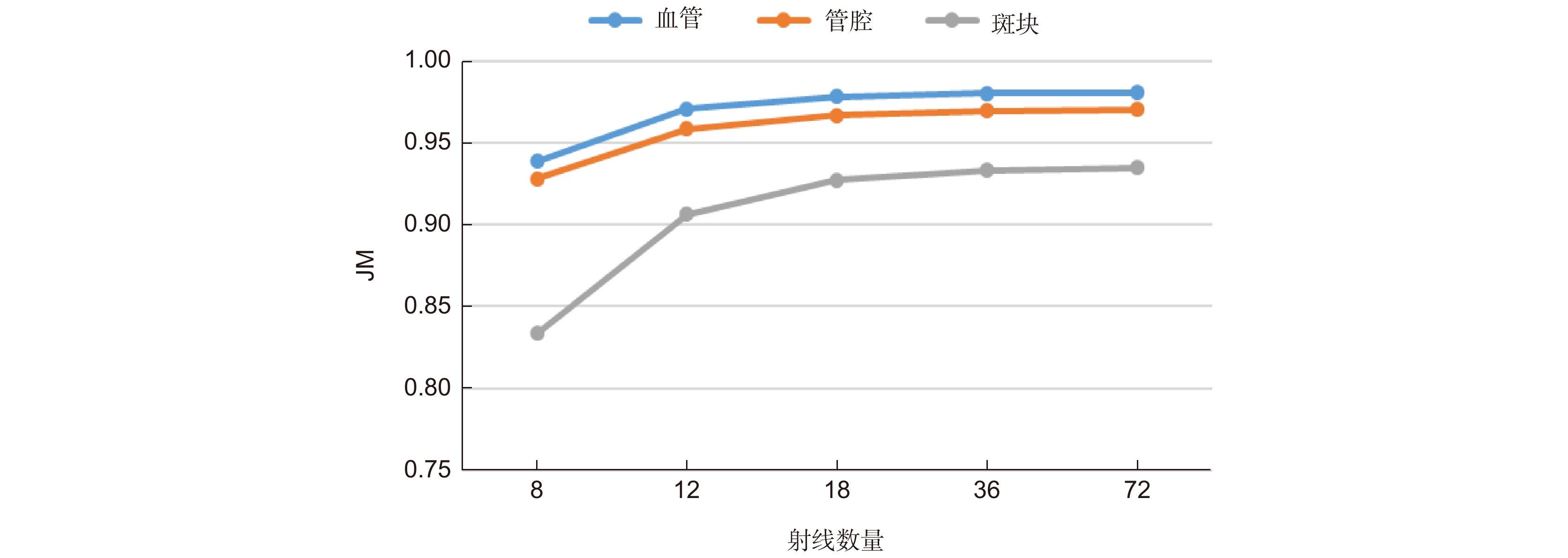
Backbone SEB num JM HD/mm PAD TER Med Lum Plaque Med Lum Med Lum - ResNet18 0 0.8630 0.8589 0.6935 0.2361 0.1501 0.1077 0.1038 0 1 0.8658 0.8520 0.6947 0.2252 0.1603 0.1048 0.1174 0 2 0.8659 0.8634 0.6979 0.2167 0.1555 0.1078 0.1039 0 3 0.8655 0.8598 0.7016 0.2258 0.1513 0.1117 0.1038 0 ResNet34 0 0.8866 0.8674 0.7302 0.1906 0.1446 0.0849 0.0950 0 1 0.8803 0.8606 0.7173 0.2080 0.1475 0.0891 0.1013 0 2 0.8716 0.8610 0.7071 0.2357 0.1558 0.0979 0.1046 0 3 0.8818 0.8574 0.7167 0.2227 0.1559 0.0833 0.1077 0 ResNet50 0 0.8804 0.8692 0.7221 0.1855 0.1462 0.0937 0.0967 0 1 0.8738 0.8687 0.7131 0.2248 0.1385 0.0978 0.0946 0 2 0.8760 0.8671 0.7152 0.2266 0.1445 0.0901 0.0990 0 3 0.8805 0.8757 0.7211 0.2153 0.1351 0.0915 0.0879 0 ResNet101 0 0.8853 0.8665 0.7298 0.2014 0.1380 0.0860 0.0928 0 1 0.8910 0.8646 0.7346 0.1873 0.1553 0.0816 0.1099 0 2 0.8934 0.8738 0.7430 0.1761 0.1355 0.0794 0.1005 0 3 0.8902 0.8725 0.7337 0.1775 0.1396 0.0745 0.0827 0 ResNet152 0 0.8852 0.8646 0.7302 0.2012 0.1412 0.0881 0.1036 0 1 0.8845 0.8711 0.7256 0.2073 0.1402 0.0874 0.0958 0 2 0.8974 0.8708 0.7424 0.1879 0.1481 0.0736 0.0962 0 3 0.8849 0.8627 0.7255 0.2101 0.1545 0.0854 0.1088 0 表 3 不同损失函数下的实验结果
Table 3. Experimental results with different loss functions
Loss function JM HD/mm PAD TER Med Lum Plaque Med Lum Med Lum - Smoothl1 0.8732 0.8698 0.7047 0.2131 0.1436 0.0999 0.0930 0 Ll+Lp 0.8850 0.8757 0.7319 0.2145 0.1350 0.0912 0.0929 0 Lm+Lp 0.8904 0.8636 0.7313 0.1997 0.1509 0.0794 0.1116 0 Ll+Lm 0.8808 0.8736 0.7183 0.2172 0.1447 0.0894 0.0892 0 IVUS Polar IoU Loss 0.8934 0.8738 0.7430 0.1761 0.1355 0.0794 0.1005 0 表 4 不同建模方式下的实验结果
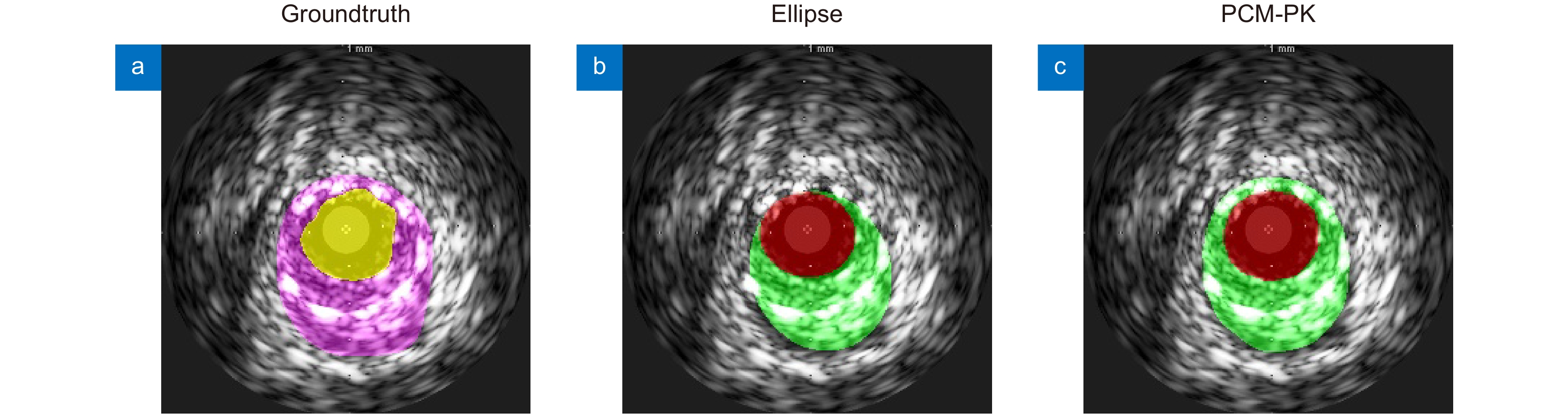
Table 4. Experimental results with different modeling methods
建模方式 Loss JM HD/mm PAD TER Med Lum Plaque Med Lum Med Lum - Ellipse Smoothl1 0.8208 0.8124 0.6100 0.2633 0.1793 0.1399 0.1402 0.0767 PCM-PK 0.8732 0.8698 0.7047 0.2131 0.1436 0.0999 0.0930 0 表 5 不同分割模型的性能比较
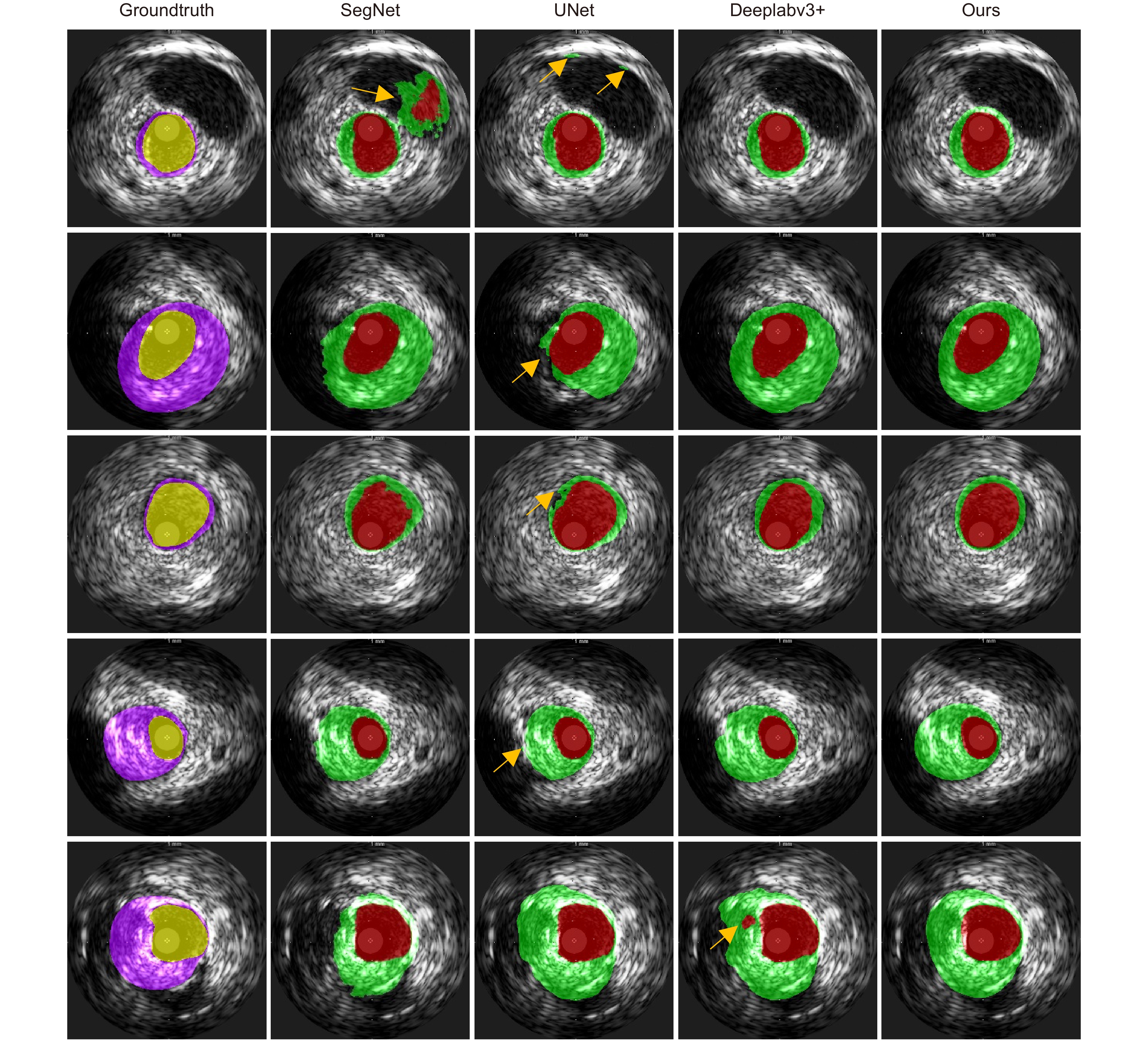
Table 5. Performance comparison of different segmentation models
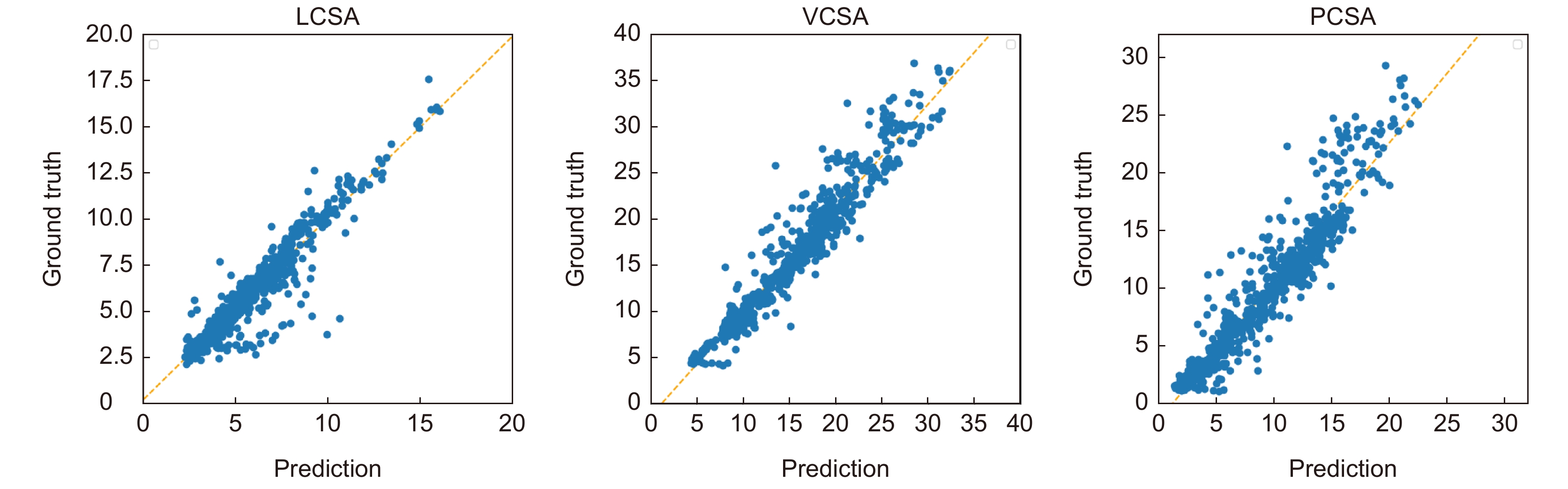
表 6 关键临床参数线性回归分析结果
Table 6. Results of linear regression analysis of key clinical parameters
斜率 截距 Pearson相关系数 LCSA 0.9825 0.2359 0.9427 VCSA 1.1259 −1.3911 0.9626 PCSA 1.2016 −1.5044 0.9432 表 7 关键临床参数Bland-Altman分析结果
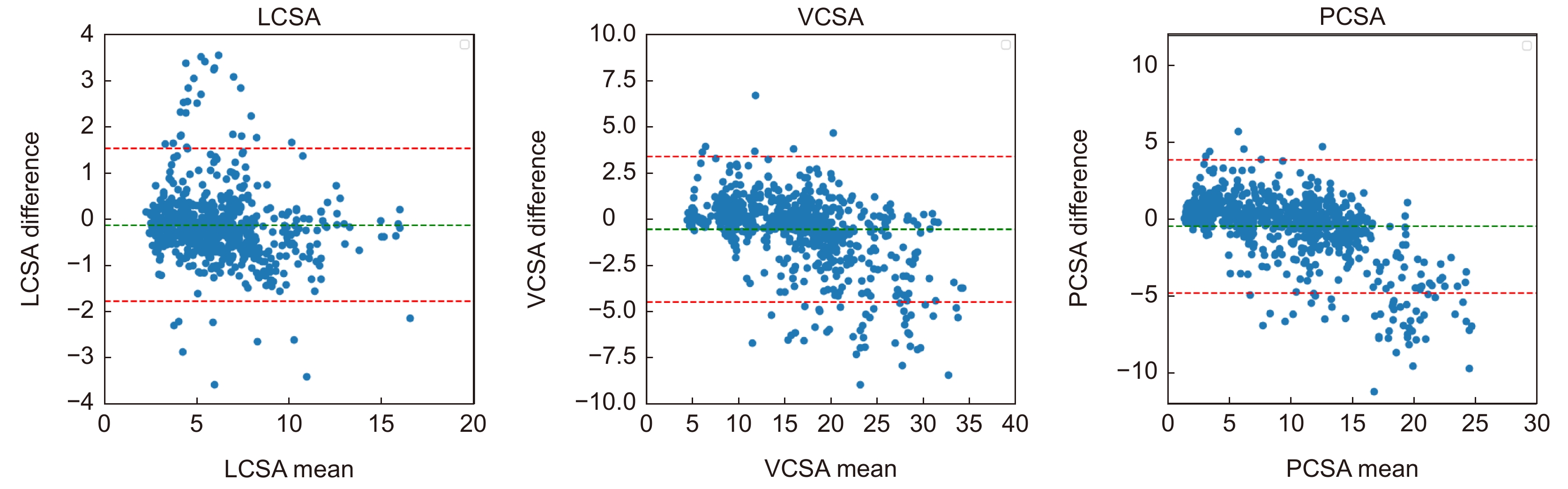
Table 7. Results of Bland-Altman analysis of key clinical parameters
均值 均值偏移 偏移程度/% 离群值比例/% LCSA 5.9898 −0.1320 −2.20 5.25 VCSA 15.8044 −0.5628 −3.56 6.59 PCSA 9.8146 −0.4308 −4.40 7.27 -
参考文献
[1] Schoenhagen P, Crowe T, Nicholls S, et al. IVUS Made Easy: An Introduction to Coronary Intravascular Ultrasound Imaging[M]. Great Britain: Informa Healthcare, 2008.
[2] Sonka M, Zhang X M, Siebes M, et al. Segmentation of intravascular ultrasound images: a knowledge-based approach[J]. IEEE Trans Med Imaging, 1995, 14(4): 719−732. doi: 10.1109/42.476113
[3] Meier D S, Cothren R M, Vince D G, et al. Automated morphometry of coronary arteries with digital image analysis of intravascular ultrasound[J]. Am Heart J, 1997, 133(6): 681−690. doi: 10.1016/S0002-8703(97)70170-4
[4] Essa E, Xie X H, Sazonov I, et al. Automatic IVUS media-adventitia border extraction using double interface graph cut segmentation[C]//18th IEEE International Conference on Image Processing, Brussels, 2011: 69−72. https://doi.org/10.1109/ICIP.2011.6116649.
[5] Kovalski G, Beyar R, Shofti R, et al. Three-dimensional automatic quantitative analysis of intravascular ultrasound images[J]. Ultrasound Med Biol, 2000, 26(4): 527−537. doi: 10.1016/S0301-5629(99)00167-2
[6] Hernandez A H, Gil D G, Radeva P R, et al. Anisotropic processing of image structures for adventitia detection in intravascular ultrasound images[C]//Computers in Cardiology, 2004, Chicago, 2004: 229−232. https://doi.org/10.1109/CIC.2004.1442914.
[7] Giannoglou G D, Chatzizisis Y S, Koutkias V, et al. A novel active contour model for fully automated segmentation of intravascular ultrasound images: in vivo validation in human coronary arteries[J]. Comput Biol Med, 2007, 37(9): 1292−1302. doi: 10.1016/j.compbiomed.2006.12.003
[8] Mendizabal-Ruiz E G, Kakadiaris I A. Probabilistic segmentation of the lumen from intravascular ultrasound radio frequency data[C]//Proceedings of the 15th International Conference on Medical Image Computing and Computer-Assisted Intervention, Nice, 2012: 454–461. https://doi.org/10.1007/978-3-642-33418-4_56.
[9] Gil D, Radeva P, Saludes J, et al. Automatic segmentation of artery wall in coronary IVUS images: a probabilistic approach[C]//Computers in Cardiology 2000, Cambridge, 2000: 687–690. https://doi.org/10.1109/CIC.2000.898617.
[10] Gil D, Radeva P, Saludes J. Segmentation of artery wall in coronary IVUS images: a probabilistic approach[C]// Proceedings of the 15th International Conference on Pattern Recognition, Barcelona, 2000: 352–355. https://doi.org/10.1109/ICPR.2000.902931.
[11] Haas C, Ermert H, Holt S, et al. Segmentation of 3D intravascular ultrasonic images based on a random field model[J]. Ultrasound Med Biol, 2000, 26(2): 297−306. doi: 10.1016/S0301-5629(99)00139-8
[12] Yang J, Tong L, Faraji M, et al. IVUS-Net: an intravascular ultrasound segmentation network[C]//Proceedings of the 1st International Conference on Smart Multimedia, Toulon, 2018: 367–377. https://doi.org/10.1007/978-3-030-04375-9_31.
[13] Yang J, Faraji M, Basu A. Robust segmentation of arterial walls in intravascular ultrasound images using Dual Path U-Net[J]. Ultrasonics, 2019, 96: 24−33. doi: 10.1016/j.ultras.2019.03.014
[14] Zhu F B, Gao Z Y, Zhao C, et al. A deep learning-based method to extract lumen and media-adventitia in intravascular ultrasound images[J]. Ultrason Imaging, 2022, 44(5−6): 191−203. doi: 10.1177/01617346221114137
[15] Kim S, Jang Y, Jeon B, et al. Fully automatic segmentation of coronary arteries based on deep neural network in intravascular ultrasound images[C]//Proceedings of the 7th Joint International Workshop, CVII-STENT 2018 and Third International Workshop, LABELS 2018, Held in Conjunction with MICCAI 2018, Granada, 2018: 161−168. https://doi.org/10.1007/978-3-030-01364-6_18.
[16] Xia M H, Yan W J, Huang Y, et al. Extracting membrane borders in IVUS images using a multi-scale feature aggregated U-Net[C]//2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Montreal, 2020: 1650−1653. https://doi.org/10.1109/EMBC44109.2020.9175970.
[17] Bajaj R, Huang X R, Kilic Y, et al. Advanced deep learning methodology for accurate, real-time segmentation of high-resolution intravascular ultrasound images[J]. Int J Cardiol, 2021, 339: 185−191. doi: 10.1016/j.ijcard.2021.06.030
[18] Blanco P J, Ziemer P G P, Bulant C A, et al. Fully automated lumen and vessel contour segmentation in intravascular ultrasound datasets[J]. Med Image Anal, 2022, 75: 102262. doi: 10.1016/j.media.2021.102262
[19] Bargsten L, Raschka S, Schlaefer A. Capsule networks for segmentation of small intravascular ultrasound image datasets[J]. Int J Comput Assist Radiol Surg, 2021, 16(8): 1243−1254. doi: 10.1007/s11548-021-02417-x
[20] Nishi T, Yamashita R, Imura S, et al. Deep learning-based intravascular ultrasound segmentation for the assessment of coronary artery disease[J]. Int J Cardiol, 2021, 333: 55−59. doi: 10.1016/j.ijcard.2021.03.020
[21] Sinha P, Wu Y M, Psaromiligkos I, et al. Lumen & media segmentation of IVUS images via ellipse fitting using a wavelet-decomposed subband CNN[C]//2020 IEEE 30th International Workshop on Machine Learning for Signal Processing (MLSP), Espoo, 2020: 1−6. https://doi.org/10.1109/MLSP49062.2020.9231871.
[22] Xie E Z, Sun P Z, Song X G, et al. PolarMask: single shot instance segmentation with polar representation[C]//2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, 2020: 12190−12199. https://doi.org/10.1109/CVPR42600.2020.01221.
[23] Schmidt U, Weigert M, Broaddus C, et al. Cell detection with star-convex polygons[C]// Proceedings of the 21st International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, 2018: 265−273. https://doi.org/10.1007/978-3-030-00934-2_30.
[24] Zhang Z L, Zhang X Y, Peng C, et al. ExFuse: enhancing feature fusion for semantic segmentation[C]//Proceedings of the 15th European Conference on Computer Vision, Munich, 2018: 273–288. https://doi.org/10.1007/978-3-030-01249-6_17.
[25] He K M, Zhang X Y, Ren S Q, et al. Deep residual learning for image recognition[C]//2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, 2016: 770–778. https://doi.org/10.1109/CVPR.2016.90.
[26] Wang H H, Wu X D, Huang Z Y, et al. High-frequency component helps explain the generalization of convolutional neural networks[C]//2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, 2020: 8681–8691. https://doi.org/10.1109/CVPR42600.2020.00871.
[27] Balocco S, Gatta C, Ciompi F, et al. Standardized evaluation methodology and reference database for evaluating IVUS image segmentation[J]. Comput Med Imaging Graph, 2014, 38(2): 70−90. doi: 10.1016/j.compmedimag.2013.07.001
[28] Wyburd M K, Dinsdale N K, Namburete A I L, et al. TEDS-Net: enforcing diffeomorphisms in spatial transformers to guarantee topology preservation in segmentations[C]//Proceedings of the 24th International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, 2021: 250–260. https://doi.org/10.1007/978-3-030-87193-2_24.
[29] Chen L C, Zhu Y K, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation[C]//Proceedings of the 15th European Conference on Computer Vision, Munich, 2018: 833‒851. https://doi.org/10.1007/978-3-030-01234-2_49.
[30] Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation[J]. IEEE Trans Pattern Anal Mach Intell, 2017, 39(12): 2481−2495. doi: 10.1109/TPAMI.2016.2644615
[31] Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation[C]. Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, 2015: 234‒241. https://doi.org/10.1007/978-3-319-24574-4_28.
-
访问统计

 E-mail Alert
E-mail Alert RSS
RSS

 下载:
下载: