-
摘要
全切片图像(Whole slide imaging, WSI)是癌症诊断和预后的关键依据,具有尺寸庞大、空间关系复杂以及风格各异等特点。由于其缺乏细节注释,传统的计算病理学方法难以处理肿瘤组织环境中的空间关系。本文提出了一种新型的基于图神经网络的WSI生存预测模型BC-GraphSurv。首先,采用迁移学习的预训练策略,构建WSI的病理关系拓扑结构,实现了对病理学图像特征和空间关系信息的有效提取。然后,采用GAT-GCN双分支结构进行预测,在图注意力网络中加入边属性和全局连接模块,同时引入图卷积网络分支补充局部细节,增强了对WSI风格差异的适应能力,能够有效利用拓扑结构处理空间关系,区分微病理环境。在WSI数据集TCGA-BRCA和TCGA-KIRC上进行的实验表明,BC-GraphSurv模型的一致性指数为0.7950和0.7458,相比于当前先进的生存预测模型提升了0.0409,充分证明了模型的有效性。
Abstract
Whole slide imaging (WSI) is the main basis for cancer diagnosis and prognosis, characterized by its large size, complex spatial relationships, and diverse styles. Due to its lack of detailed annotations, traditional computational pathology methods are difficult to handle WSI tasks. To address these challenges, this paper proposes a WSI survival prediction model based on graph neural networks, BC-GraphSurv. Specifically, we use transfer learning pre-training to extract features containing spatial relationship information and construct the pathological relationship topology of WSI. Then, the two branch structures of the improved graph attention network (GAT) and graph convolution network (GCN) are used to predict the extracted features. We combine edge attributes and global perception modules in GAT, while the GCN branch is used to supplement local details, which can achieve adaptability to WSI style differences and effectively utilize topological structures to handle spatial relationships and distinguish subtle pathological environments. Experimental results on the TCGA-BRCA dataset demonstrate BC-GraphSurv's effectiveness, achieving a C-index of 0.795—a significant improvement of 0.0409 compared to current state-of-the-art survival prediction models. This underscores its robust efficacy in addressing WSI challenges in cancer diagnosis and prognosis.
-
Key words:
- survival prediction /
- graph neural network /
- transfer learning /
- attention mechanism
-
Overview
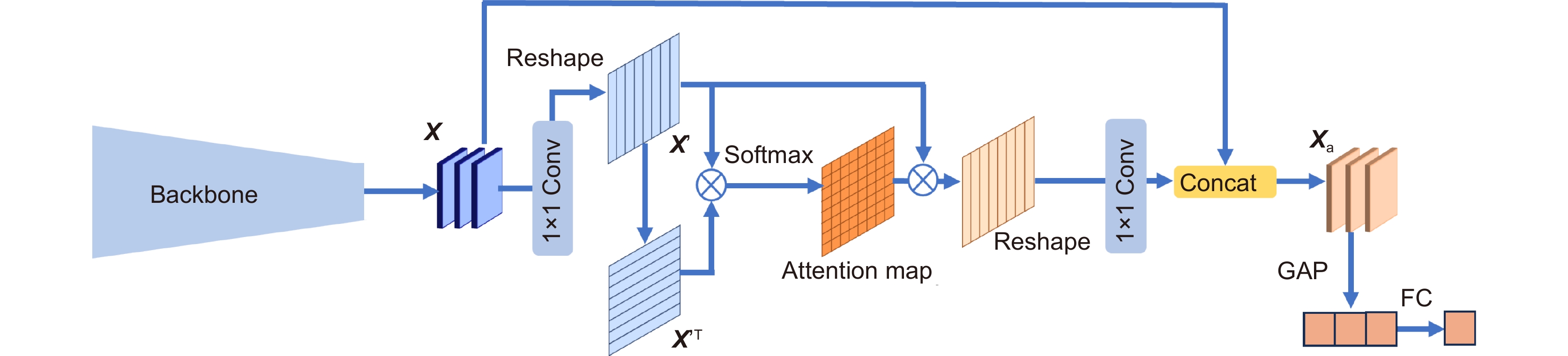
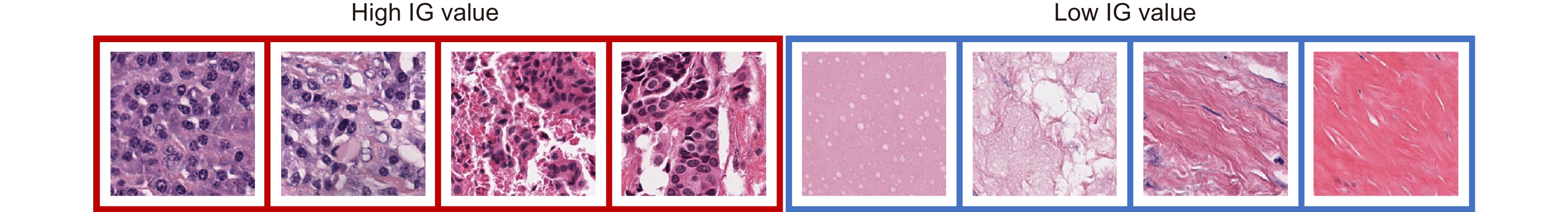
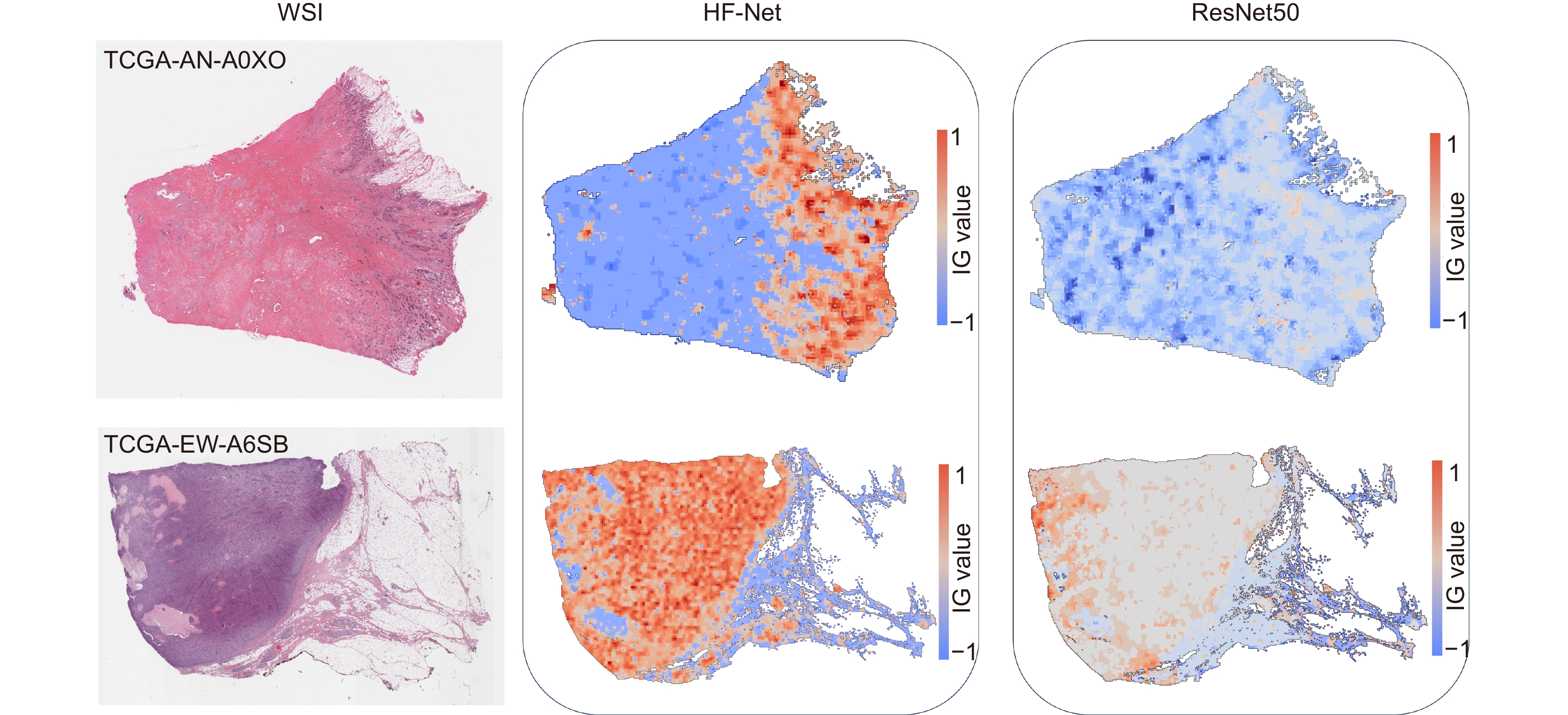
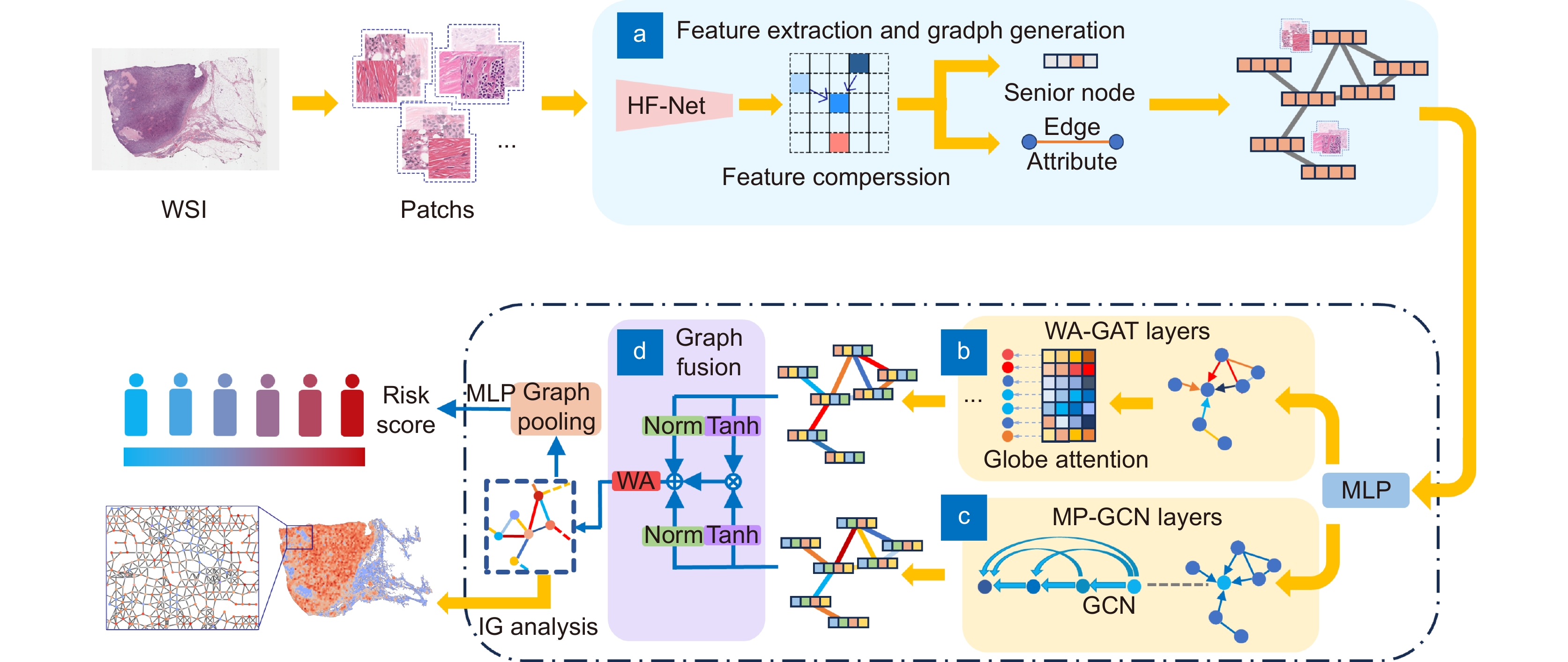
Overview: In this study, we present BC-GraphSurv, an innovative model for breast cancer survival prediction utilizing Whole Slide Imaging (WSI). Given the challenges of large size, complex spatial relationships, and diverse styles in WSIs, BC-GraphSurv addresses these issues through a novel approach that integrates transfer learning and feature extraction using the HF-Net. The model consists of four steps: transfer learning with HF-Net, compression and fusion of similar features, construction of graph structure features, and learning with WA-GAT and MP-GCN. The model commences with a transfer learning pre-training strategy, utilizing HF-Net to construct the pathological relationship topology of WSIs. This strategy facilitates the effective extraction of features and spatial relationship information. HF-Net, trained on a breast cancer tumor classification dataset, is crucial for adapting a general backbone network to the complexity of tumor structures and tissue texture features. This network reduces noise in non-cancerous regions and enhances differentiation between cancerous and non-cancerous areas. The feature extraction network, combining Convolutional Neural Networks (CNN) and self-attention mechanisms, benefits from transfer learning to enhance pathology feature recognition via a feature transfer module. This module, coupled with spatial correlation and semantic similarity integration, enables compressed graph modeling and extraction of crucial contextual features for survival prediction. To overcome specific challenges in WSI tasks, BC-GraphSurv introduces improvements to the Graph Attention Network (GAT) in the form of the Whole Association Graph Attention Network (WA-GAT). This prediction branch employs cross-attention on node and edge features, a global perception module, and a Dense Graph Convolutional Network (GCN) for fine-grained details. The integration of WA-GAT and GCN enhances the model's adaptability to diverse WSI styles and spatial differences, effectively processing spatial information and improving analytical capabilities. Experimental validation involves ablation experiments assessing the impact of different modules and improvements. Comparative experiments with various models and visual analyses confirm the effectiveness of BC-GraphSurv. In conclusion, BC-GraphSurv provides a comprehensive solution for breast cancer survival prediction using WSIs. Experimental results on the TCGA-BRCA dataset showcase its effectiveness, with a consistency index of 0.795, surpassing current state-of-the-art models. The model's innovations effectively tackle the challenges inherent in WSI survival prediction, demonstrating robustness and superiority.
-
-
图 1 BC-GraphSurv模型架构,主要包括WSI预处理,以及(a)特征提取和生成图结构,(b) WA-GAT分支,(c) MP-GCN分支、(d)特征融合等模块
Figure 1. Architecture of the BC GraphSurv model mainly including modules such as WSI preprocessing, (a) Feature extraction and graph structure generation, (b) WA-GAT branch, (c) MP-GCN branches, and (d) Feature fusion
表 1 特征提取网络实验结果对比
Table 1. Comparison of experimental results of feature extraction network
C-index SD Parameters/MB ResNet50 0.7225 0.011 97.49 EfficientNet-b5 0.7306 0.022 115.93 HF-Net 0.7950 0.013 113.72 表 2 消融实验结果对比
Table 2. Comparison of ablation experiment results
WA-GAT MP-GCN GAT GCN GF C-index SD √ 0.7506 0.021 √ 0.7217 0.019 √ 0.7842 0.007 √ 0.7812 0.008 √ √ 0.7910 0.003 √ √ √ 0.7950 0.013 表 3 不同算法对比试验1
Table 3. First comparison experiments of different methods
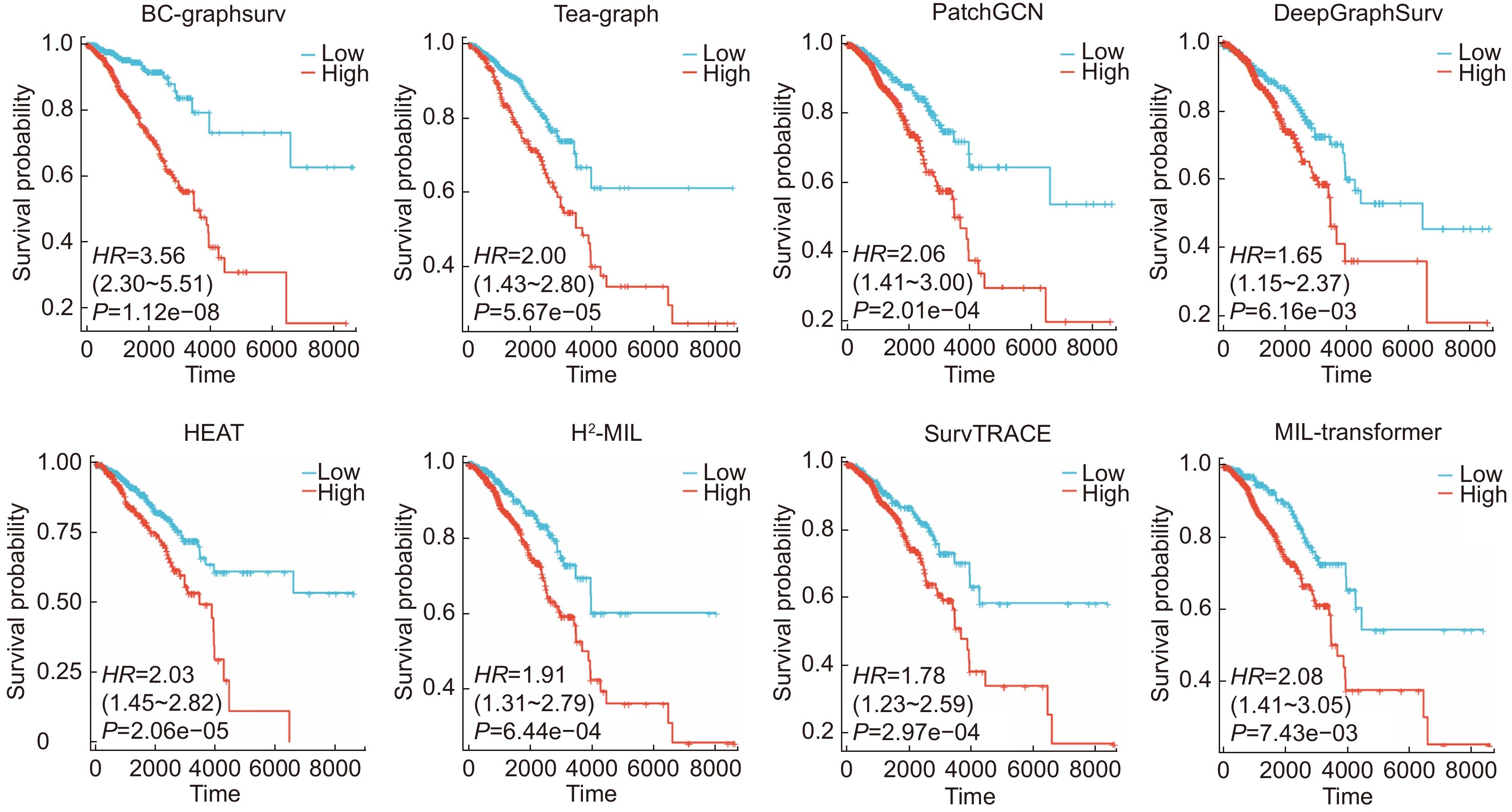
Method BRCA KIRC C-index SD C-index SD MLP 0.6176 0.027 0.5862 0.019 Attention-MIL[26] (2018) 0.7091 0.052 0.6590 0.044 WSISA[8] (2017) 0.6802 0.083 0.6151 0.057 DeepGraphSurv[12] (2018) 0.7402 0.012 0.6945 0.045 Patch GCN[13] (2021) 0.7514 0.036 0.6775 0.067 H2-MIL[27] (2022) 0.7338 0.055 0.6915 0.024 Tea-Graph[28] (2022) 0.7541 0.021 0.7109 0.023 HEAT[3] (2023) 0.7529 0.009 0.7011 0.018 BC-GraphSurv 0.7950 0.013 0.7458 0.020 -
参考文献
[1] Li X T, Li C, Rahaman M M, et al. A comprehensive review of computer-aided whole-slide image analysis: from datasets to feature extraction, segmentation, classification and detection approaches[J]. Artif Intell Rev, 2022, 55(6): 4809−4878. doi: 10.1007/s10462-021-10121-0
[2] 黄盼, 何鹏, 杨兴, 等. 基于自适应融合和显微成像的乳腺肿瘤分级网络[J]. 光电工程, 2023, 50(1): 220158. doi: 10.12086/oee.2023.220158
Huang P, He P, Yang X, et al. Breast tumor grading network based on adaptive fusion and microscopic imaging[J]. Opto-Electron Eng, 2023, 50(1): 220158. doi: 10.12086/oee.2023.220158
[3] Chan T H, Cendra F J, Ma L, et al. Histopathology whole slide image analysis with heterogeneous graph representation learning[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, Canada, 2023: 15661–15670. https://doi.org/10.1109/CVPR52729.2023.01503.
[4] 梁礼明, 卢宝贺, 龙鹏威, 等. 自适应特征融合级联Transformer视网膜血管分割算法[J]. 光电工程, 2023, 50(10): 230161. doi: 10.12086/oee.2023.230161
Liang L M, Lu B H, Long P W, et al. Adaptive feature fusion cascade Transformer retinal vessel segmentation algorithm[J]. Opto-Electron Eng, 2023, 50(10): 230161. doi: 10.12086/oee.2023.230161
[5] 吕佳, 王泽宇, 梁浩城. 边界注意力辅助的动态图卷积视网膜血管分割[J]. 光电工程, 2023, 50(1): 220116. doi: 10.12086/oee.2023.220116
Lv J, Wang Z Y, Liang H C. Boundary attention assisted dynamic graph convolution for retinal vascular segmentation[J]. Opto-Electron Eng, 2023, 50(1): 220116. doi: 10.12086/oee.2023.220116
[6] Tellez D, Litjens G, van der Laak J, et al. Neural image compression for gigapixel histopathology image analysis[J]. IEEE Trans Pattern Anal Mach Intell, 2021, 43(2): 567−578. doi: 10.1109/TPAMI.2019.2936841
[7] Zhu X L, Yao J W, Huang J Z. Deep convolutional neural network for survival analysis with pathological images[C]//2016 IEEE International Conference on Bioinformatics and Biomedicine, Shenzhen, China, 2016: 544–547. https://doi.org/10.1109/BIBM.2016.7822579.
[8] Zhu X L, Yao J W, Zhu F Y, et al. WSISA: making survival prediction from whole slide histopathological images[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 2017: 6855–6863. https://doi.org/10.1109/CVPR.2017.725.
[9] Yao J W, Zhu X L, Jonnagaddala J, et al. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks[J]. Med Image Anal, 2020, 65: 101789. doi: 10.1016/j.media.2020.101789
[10] Fan L, Sowmya A, Meijering E, et al. Cancer survival prediction from whole slide images with self-supervised learning and slide consistency[J]. IEEE Trans Med Imaging, 2023, 42(5): 1401−1412. doi: 10.1109/TMI.2022.3228275
[11] Kim H E, Cosa-Linan A, Santhanam N, et al. Transfer learning for medical image classification: a literature review[J]. BMC Med Imaging, 2022, 22(1): 69. doi: 10.1186/s12880-022-00793-7
[12] Li R Y, Yao J W, Zhu X L, et al. Graph CNN for survival analysis on whole slide pathological images[C]//21st International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 2018: 174–182. https://doi.org/10.1007/978-3-030-00934-2_20.
[13] Chen R J, Lu M Y, Shaban M, et al. Whole slide images are 2D point clouds: context-aware survival prediction using patch-based graph convolutional networks[C]//24th International Conference on Medical Image Computing and Computer Assisted Intervention, Strasbourg, France, 2021: 339–349. https://doi.org/10.1007/978-3-030-87237-3_33.
[14] Di D L, Zhang J, Lei F Q, et al. Big-Hypergraph factorization neural network for survival prediction from whole slide image[J]. IEEE Trans Image Process, 2022, 31: 1149−1160. doi: 10.1109/TIP.2021.3139229
[15] Velickovic P, Cucurull G, Casanova A, et al. Graph attention networks[C]//6th International Conference on Learning Representations, Vancouver, Canada, 2018: 1–12.
[16] Ozyoruk K B, Can S, Darbaz B, et al. A deep-learning model for transforming the style of tissue images from cryosectioned to formalin-fixed and paraffin-embedded[J]. Nat Biomed Eng, 2022, 6(12): 1407−1419. doi: 10.1038/s41551-022-00952-9
[17] Kipf T N, Welling M. Semi-supervised classification with graph convolutional networks[C]//5th International Conference on Learning Representations, Toulon, France, 2017: 1–14.
[18] Gao Z Y, Lu Z Y, Wang J, et al. A convolutional neural network and graph convolutional network based framework for classification of breast histopathological images[J]. IEEE J Biomed Health Inform, 2022, 26(7): 3163−3173. doi: 10.1109/JBHI.2022.3153671
[19] Katzman J L, Shaham U, Cloninger A, et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network[J]. BMC Med Res Methodol, 2018, 18(1): 24. doi: 10.1186/s12874-018-0482-1
[20] Kandoth C, McLellan M D, Vandin F, et al. Mutational landscape and significance across 12 major cancer types[J]. Nature, 2013, 502(7471): 333−339. doi: 10.1038/nature12634
[21] Spanhol F A, Oliveira L S, Petitjean C, et al. A dataset for breast cancer histopathological image classification[J]. IEEE Trans Biomed Eng, 2016, 63(7): 1455−1462. doi: 10.1109/TBME.2015.2496264
[22] Otsu N. A threshold selection method from gray-level histograms[J]. IEEE Trans Syst Man Cybern, 1979, 9(1): 62−66. doi: 10.1109/TSMC.1979.4310076
[23] Heagerty P J, Zheng Y Y. Survival model predictive accuracy and ROC curves[J]. Biometrics, 2005, 61(1): 92−105. doi: 10.1111/j.0006-341X.2005.030814.x
[24] Loshchilov I, Hutter F. Decoupled weight decay regularization[C]//7th International Conference on Learning Representations, New Orleans, LA, USA, 2019: 1–8.
[25] Smith L N, Topin N. Super-convergence: very fast training of neural networks using large learning rates[J]. Proc SPIE, 2019, 11006: 1100612. doi: 10.1117/12.2520589
[26] Ilse M, Tomczak J, Welling M. Attention-based deep multiple instance learning[C]//35th International Conference on Machine Learning, Stockholm, Sweden, 2018: 2127–2136.
[27] Hou W T, Yu L Q, Lin C X, et al. H2-MIL: exploring hierarchical representation with heterogeneous multiple instance learning for whole slide image analysis[C]//Proceedings of the 36th AAAI Conference on Artificial Intelligence, 2022: 933–941. https://doi.org/10.1609/aaai.v36i1.19976.
[28] Lee Y, Park J H, Oh S, et al. Derivation of prognostic contextual histopathological features from whole-slide images of tumours via graph deep learning[J]. Nat Biomed Eng, 2022. https://doi.org/10.1038/s41551-022-00923-0.
[29] Chen R J, Lu M Y, Weng W H, et al. Multimodal co-attention transformer for survival prediction in gigapixel whole slide images[C]//Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 2021: 3995–4005. https://doi.org/10.1109/ICCV48922.2021.00398.
[30] Wang Z F, Sun J M. SurvTRACE: transformers for survival analysis with competing events[C]//Proceedings of the 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Northbrook, IL, USA, 2022: 49. https://doi.org/10.1145/3535508.3545521.
[31] Lakatos E. Sample sizes based on the log-rank statistic in complex clinical trials[J]. Biometrics, 1988, 44(1): 229−241. doi: 10.2307/2531910
-
访问统计

 E-mail Alert
E-mail Alert RSS
RSS
 下载:
下载: