-
摘要
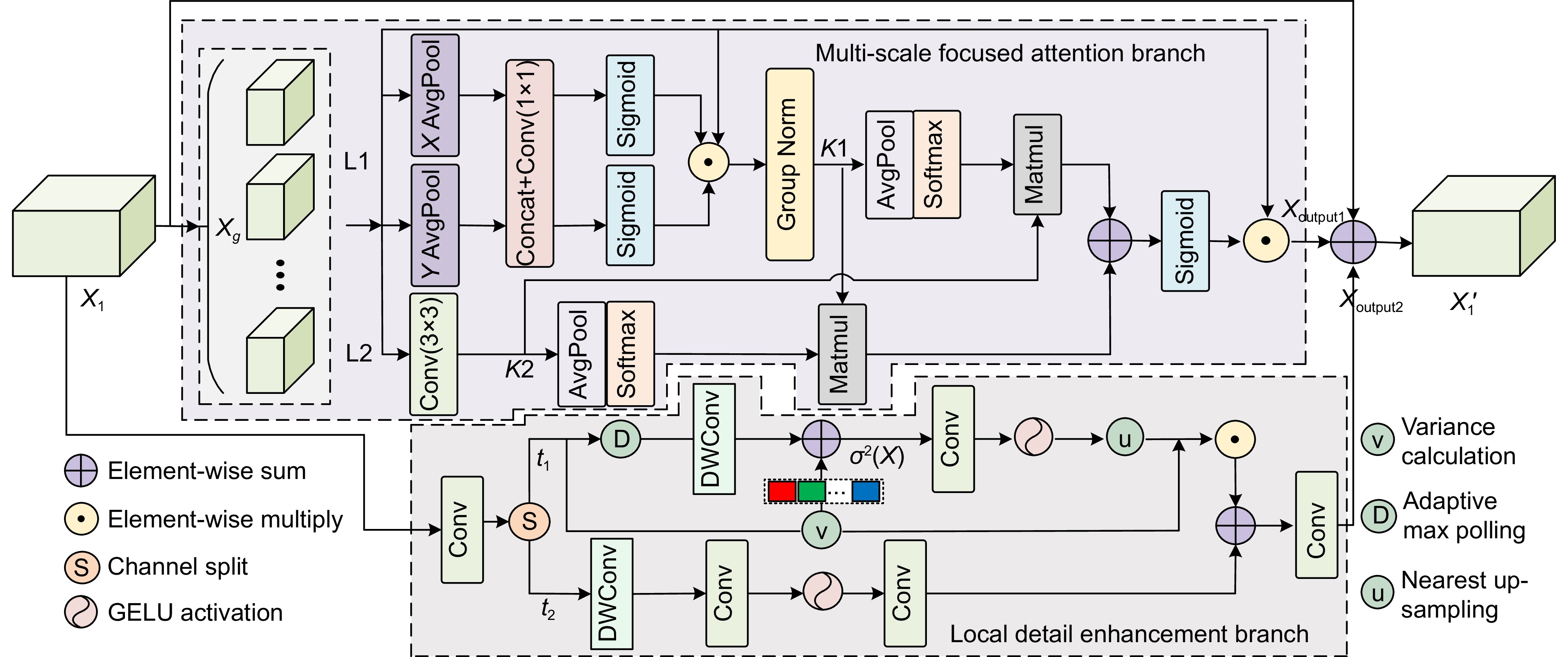
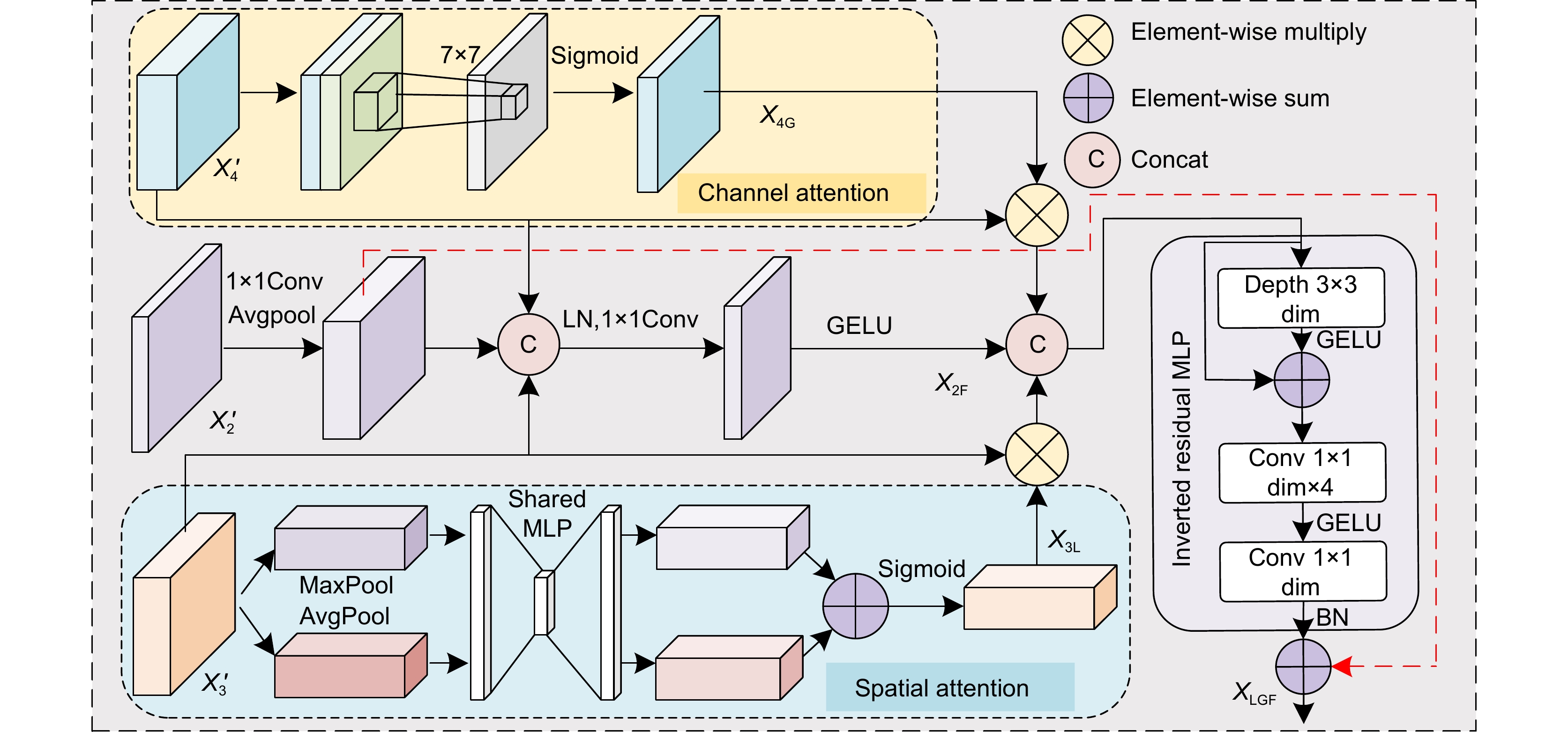
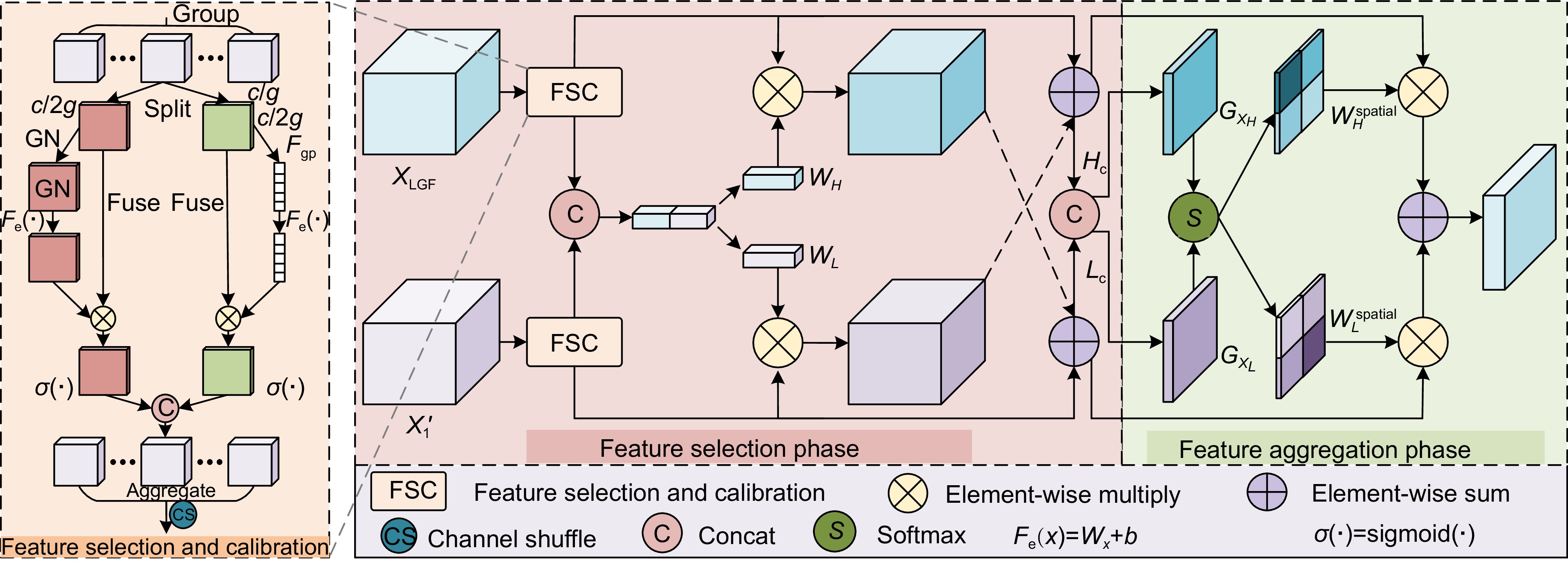
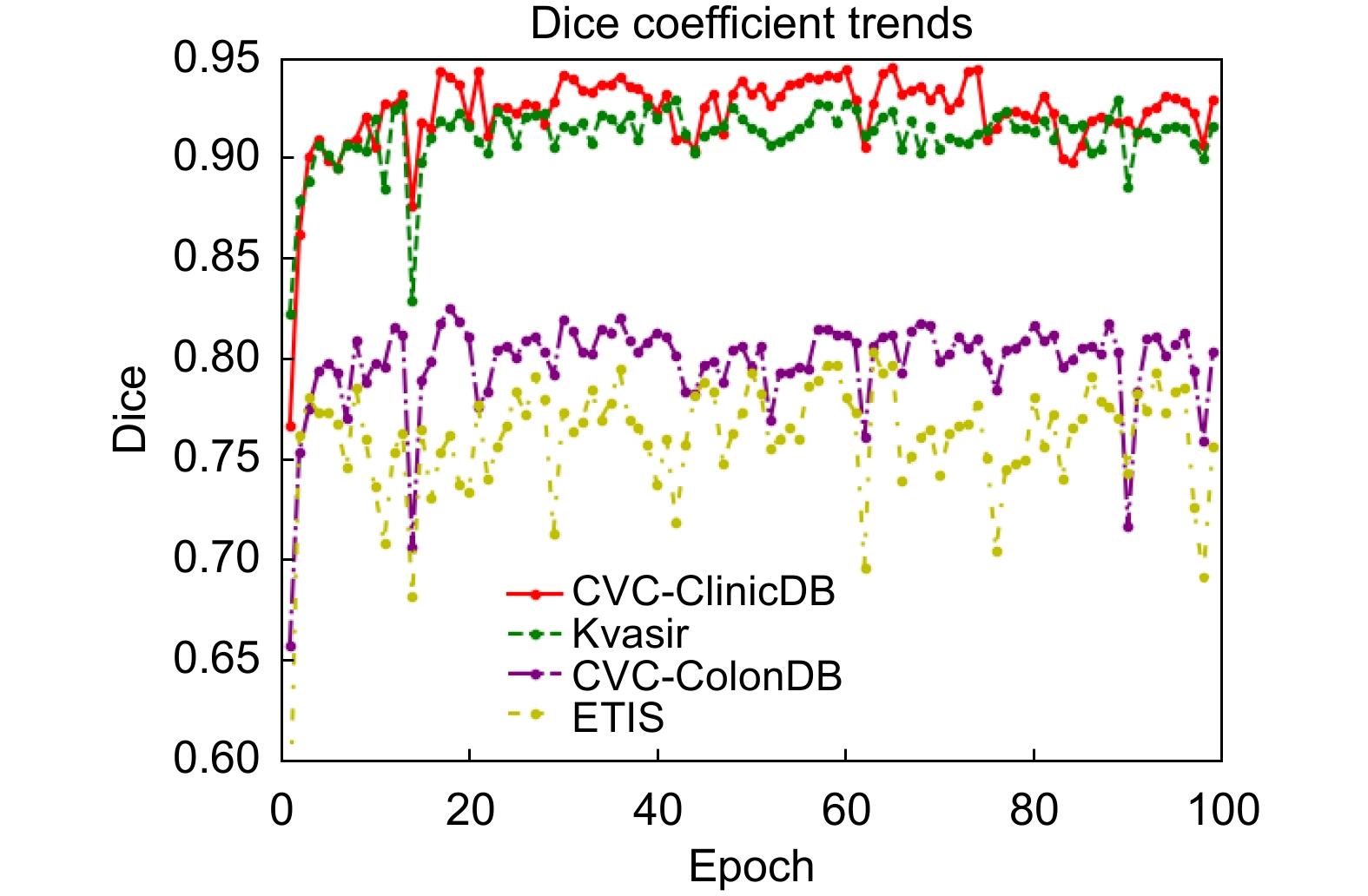
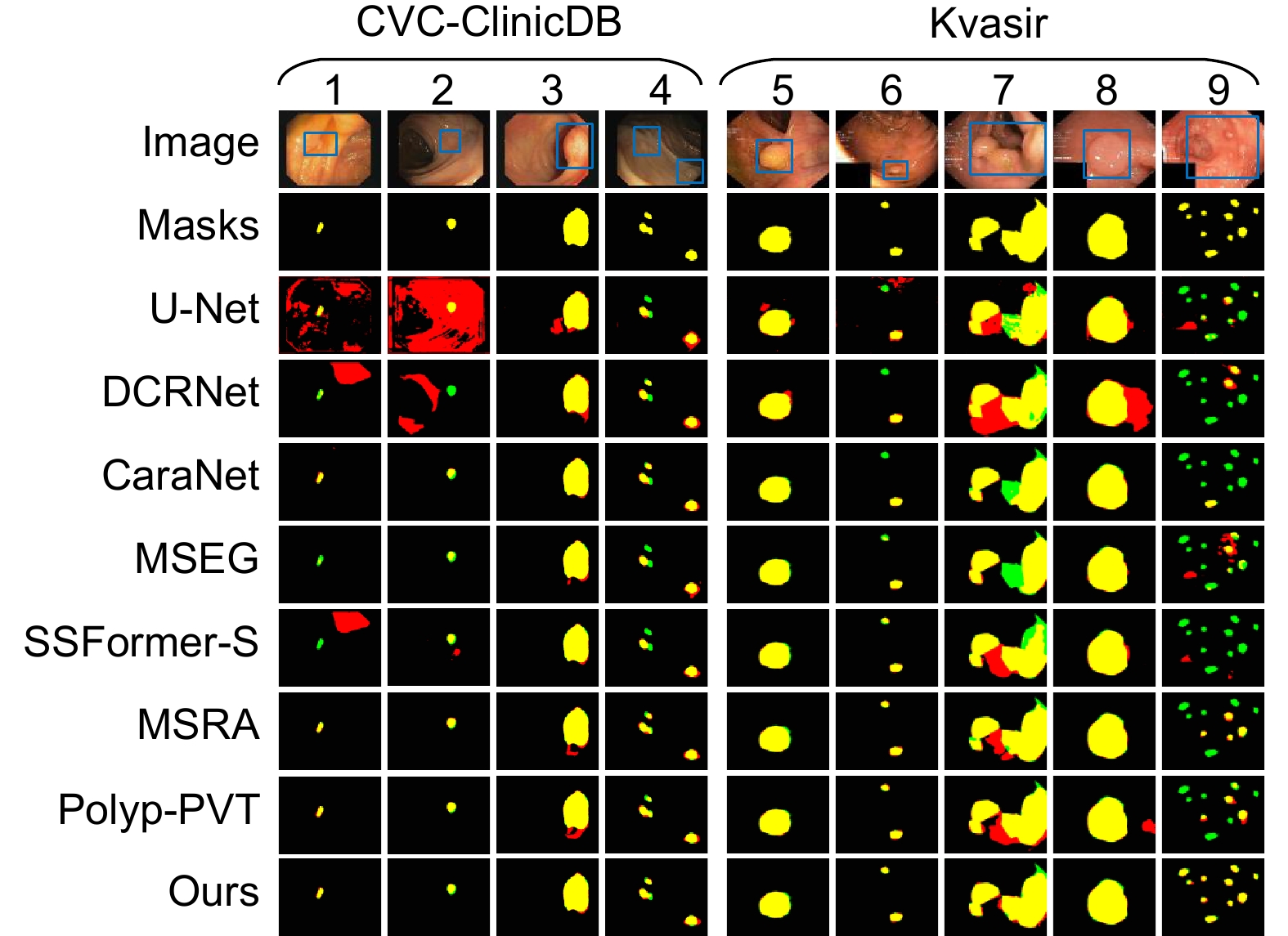
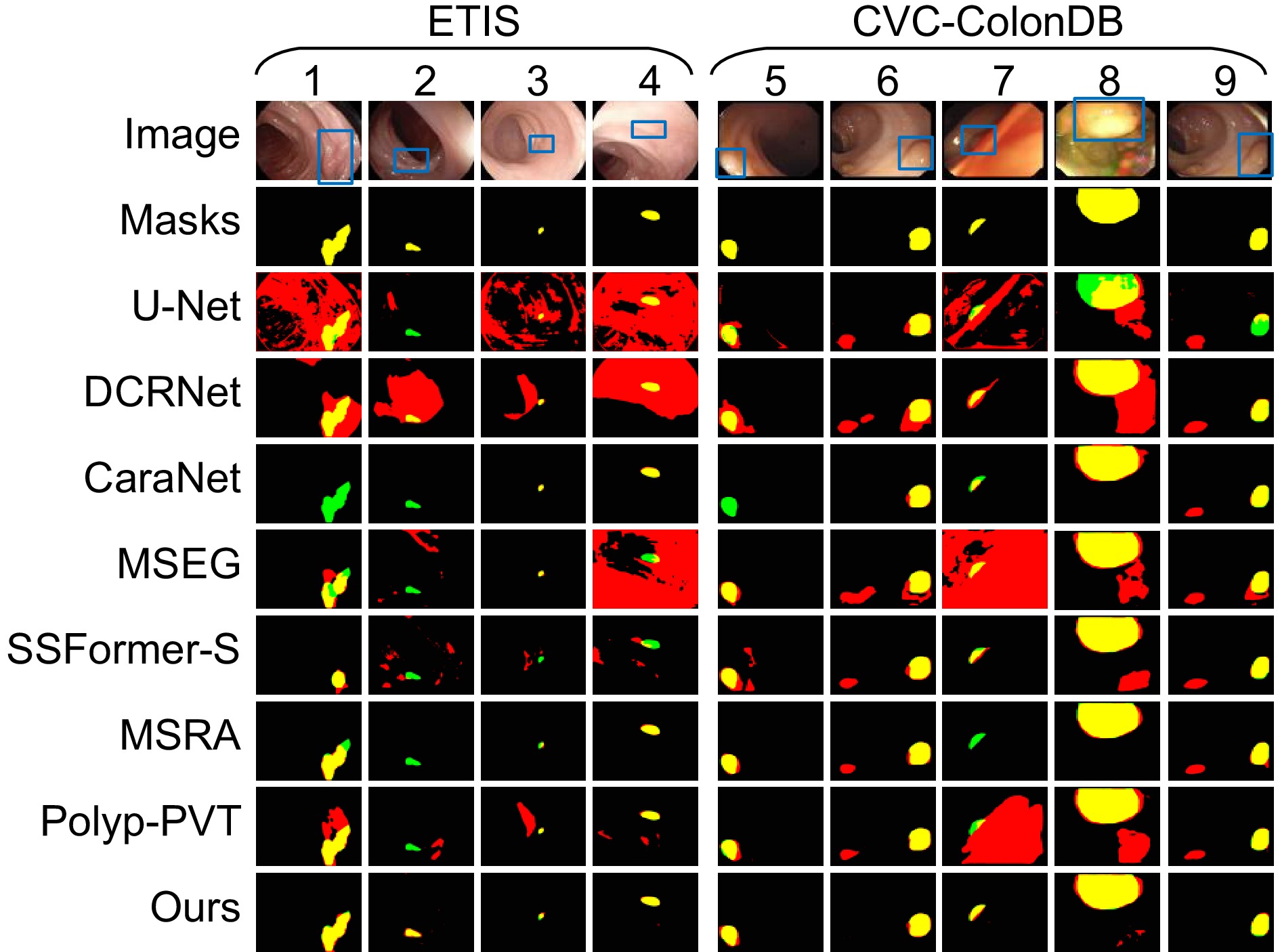
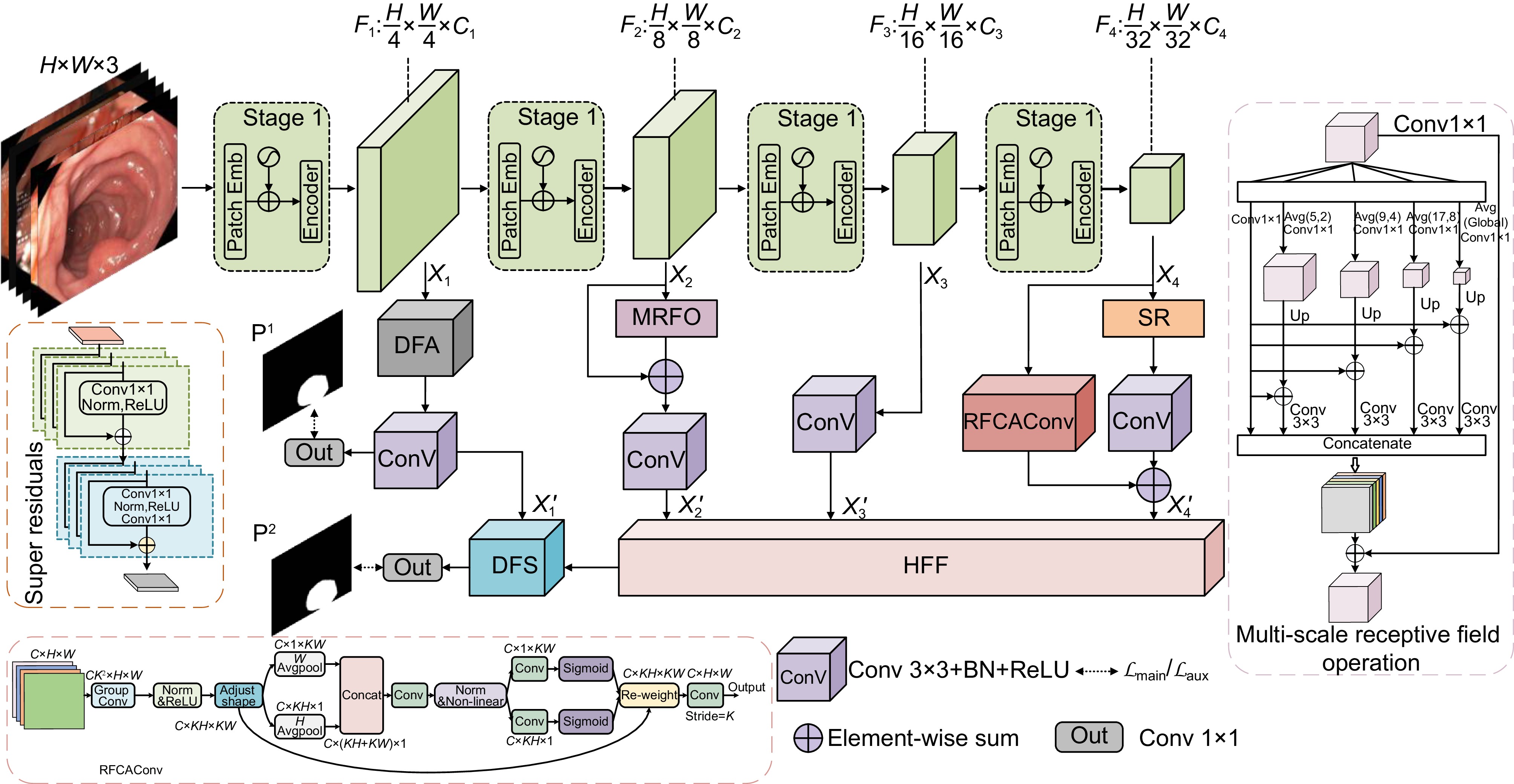
针对结直肠息肉分割中区域误分割和目标定位精度不足等挑战,本文提出一种融合Transformer自适应特征选择的结直肠息肉分割算法。首先通过Transformer编码器提取多层次特征表示,涵盖从细粒度到高层语义的多尺度信息;其次设计双重聚焦注意力模块,通过融合多尺度信息、空间注意力和局部细节特征,增强特征表达与辨识能力,显著提升病灶区域定位精度;再次引入分层特征融合模块,采用层次化聚合策略,加强局部与全局特征的融合,强化对复杂区域特征的捕捉,有效减少误分割现象;最后结合动态特征选择模块的自适应筛选与加权机制,优化多分辨率特征表达,去除冗余信息,聚焦关键区域。在Kvasir、CVC-ClinicDB、CVC-ColonDB和ETIS数据集上进行实验验证,其Dice系数分别达到0.926、0.941、0.814和0.797。实验结果表明,本文算法在结直肠息肉分割任务中具有优越性能和应用价值。
-
关键词:
- 结直肠息肉 /
- Transformer /
- 双重聚焦注意力模块 /
- 动态特征选择模块
Abstract
To address challenges such as regional mis-segmentation and insufficient target localization accuracy in colorectal polyp segmentation, this paper proposes a colorectal polyp segmentation algorithm that integrates adaptive feature selection based on a Transformer. Firstly, the Transformer encoder is employed to extract multi-level feature representations, capturing multi-scale information from fine-grained to high-level semantics. Secondly, a dual-focus attention module is designed to enhance feature representation and recognition capabilities by integrating multi-scale information, spatial attention, and local detail features, significantly improving the localization accuracy of lesion areas. Thirdly, a hierarchical feature fusion module is introduced, which adopts a hierarchical aggregation strategy to strengthen the fusion of local and global features, enhancing the capture of complex regional features and effectively reducing mis-segmentation. Finally, a dynamic feature selection module is incorporated with adaptive selection and weighting mechanisms to optimize multi-resolution feature representation, eliminate redundant information, and focus on key areas. Experiments conducted on the Kvasir, CVC-ClinicDB, CVC-ColonDB, and ETIS datasets achieved Dice coefficients of 0.926, 0.941, 0.814, and 0.797, respectively. The experimental results demonstrate that the proposed algorithm exhibits superior performance and application value in the task of colorectal polyp segmentation.
-
Overview
Overview: Colorectal cancer ranks among the most common and life-threatening diseases worldwide, with colorectal polyps identified as the primary precursors. Accurate detection and segmentation of polyps are essential for preventing cancer progression and improving patient outcomes. However, existing segmentation methods face persistent challenges, including regional mis-segmentation, low localization accuracy, and difficulties in capturing the complex features of polyps. To overcome these limitations, this study presents a novel colorectal polyp segmentation algorithm that integrates Transformer-based adaptive feature selection to improve segmentation accuracy and robustness.
The proposed approach utilizes a Transformer encoder to extract multi-level feature representations, capturing information from fine-grained details to high-level semantics. This enables a comprehensive understanding of the morphology of polyps and their surrounding tissues. To further improve feature representation, a dual-focus attention module is introduced, which integrates multi-scale information, spatial attention, and local detail features. This module enhances lesion localization accuracy and reduces errors arising from the complex structures of polyps.
To address regional mis-segmentation, a hierarchical feature fusion module is developed. By employing a hierarchical aggregation strategy, this module strengthens the integration of local and global features, allowing the model to better capture intricate regional characteristics. Additionally, a dynamic feature selection module is incorporated to optimize multi-resolution feature representations. Through adaptive selection and weighting mechanisms, this module eliminates redundant information and focuses on critical regions, improving segmentation precision.
In conclusion, this study demonstrates the effectiveness of integrating Transformer-based adaptive feature selection, dual-focus attention, hierarchical feature fusion, and dynamic feature optimization. The proposed algorithm provides a comprehensive and innovative solution to the challenges of colorectal polyp segmentation, offering significant potential for clinical applications in early cancer diagnosis and treatment.
-
-
表 1 数据集细节及划分
Table 1. Dataset details and division
Dataset Image resolution Train data Test data Image type CVC-ClinicDB 384×288 550 62 Image Kvasir-SEG Size variation 900 100 Image and mask CVC-ColonDB 574×500 0 380 Image ETIS 1226×996 0 196 Image 表 2 Kvasir和CVC-ClinicDB数据集上不同网络分割结果
Table 2. Segmentation results of different networks on Kvasir and CVC-ClinicDB datasets
Dataset Method Dice MIoU SE PC F2 MAE Kvasir U-Net[4] 0.818 0.746 0.856 0.857 0.827 0.055 DCRNet[6] 0.888 0.825 0.902 0.904 0.891 0.035 CaraNet[7] 0.922 0.872 0.915 0.941 0.921 0.019 MSEG[8] 0.899 0.842 0.900 0.923 0.896 0.028 SSFormer-S[9] 0.925 0.877 0.914 0.944 0.917 0.018 MSRAFormer[10] 0.923 0.873 0.915 0.952 0.917 0.024 Polyp-PVT[21] 0.917 0.864 0.913 0.947 0.914 0.023 Ours 0.926 0.879 0.917 0.955 0.919 0.023 CVC-ClinicDB U-Net[4] 0.823 0.755 0.834 0.839 0.827 0.019 DCRNet[6] 0.899 0.846 0.913 0.893 0.906 0.010 CaraNet[7] 0.934 0.890 0.944 0.940 0.939 0.006 MSEG[8] 0.912 0.866 0.924 0.935 0.918 0.007 SSFormer-S[9] 0.918 0.875 0.905 0.939 0.910 0.007 MSRAFormer[10] 0.924 0.874 0.945 0.920 0.932 0.008 Polyp-PVT[21] 0.937 0.889 0.949 0.936 0.945 0.006 Ours 0.941 0.896 0.957 0.934 0.949 0.006 表 3 CVC-ConlonDB和ETIS数据集上不同网络分割结果
Table 3. Segmentation results of different networks on CVC-ColonDB and ETIS datasets
Dataset Method Dice MIoU SE PC F2 MAE CVC-ConlonDB U-Net[4] 0.512 0.444 0.523 0.621 0.510 0.061 DCRNet[6] 0.707 0.632 0.776 0.719 0.723 0.052 CaraNet[7] 0.748 0.683 0.753 0.893 0.746 0.035 MSEG[8] 0.738 0.669 0.752 0.806 0.739 0.038 SSFormer-S[9] 0.774 0.698 0.777 0.837 0.766 0.036 MSRAFormer[10] 0.782 0.707 0.803 0.874 0.181 0.028 Polyp-PVT[21] 0.808 0.727 0.821 0.849 0.809 0.031 Ours 0.814 0.732 0.849 0.824 0.825 0.028 ETIS U-Net[4] 0.398 0.335 0.482 0.439 0.429 0.036 DCRNet[6] 0.550 0.486 0.746 0.504 0.600 0.095 CaraNet[7] 0.728 0.661 0.775 0.814 0.750 0.017 MSEG[8] 0.703 0.632 0.739 0.710 0.720 0.015 SSFormer-S[9] 0.769 0.698 0.856 0.743 0.800 0.016 MSRAFormer[10] 0.750 0.679 0.811 0.745 0.777 0.013 Polyp-PVT[21] 0.787 0.706 0.867 0.774 0.820 0.013 Ours 0.797 0.716 0.889 0.761 0.834 0.018 表 4 不同网络性能对比(CVC-ClinicDB)
Table 4. Performance comparison of different networks (CVC-ClinicDB)
Method Parameters/M GFLOPs Train/($ {\text{round}} \cdot {{\text{s}}^{ - 1}} $) U-Net 34.53 65.52 309 DCRNet 28.70 53.00 285 CaraNet 44.54 11.45 256 SSFormer-S 29.31 10.11 220 MSRAFormer 68.96 21.29 199 Polyp-PVT 25.12 5.30 233 Ours 26.05 11.00 183 表 5 各模块在CVC-ClinicDB数据集上的消融研究结果
Table 5. Ablation results of each module on the CVC-ClinicDB dataset
Method DFA HFF DFS Dice MIoU SE PC F2 G1 √ √ 0.929 0.882 0.935 0.941 0.931 G2 √ √ 0.930 0.884 0.950 0.924 0.938 G3 √ √ 0.921 0.866 0.922 0.935 0.921 G4 √ √ √ 0.941 0.896 0.957 0.934 0.949 表 6 各模块在ETIS数据集上的消融研究结果
Table 6. Ablation results of each module on the ETIS dataset
Method DFA HFF DFS Dice MIoU SE PC F2 G1 √ √ 0.782 0.705 0.856 0.748 0.816 G2 √ √ 0.787 0.705 0.875 0.755 0.825 G3 √ √ 0.780 0.698 0.839 0.771 0.806 G4 √ √ √ 0.797 0.716 0.889 0.761 0.834 -
参考文献
[1] 谢斌, 刘阳倩, 李俞霖. 结合极化自注意力和Transformer的结直肠息肉分割方法[J]. 光电工程, 2024, 51(10): 240179. doi: 10.12086/oee.2024.240179
Xie B, Liu Y Q, Li Y L. Colorectal polyp segmentation method combining polarized self-attention and Transformer[J]. Opto-Electron Eng, 2024, 51(10): 240179. doi: 10.12086/oee.2024.240179
[2] Lin L, Lv G Z, Wang B, et al. Polyp-LVT: polyp segmentation with lightweight vision transformers[J]. Knowledge-Based Syst, 2024, 300: 112181. doi: 10.1016/j.knosys.2024.112181
[3] 张艳, 马春明, 刘树东, 等. 基于多尺度特征增强的高效Transformer语义分割网络[J]. 光电工程, 2024, 51(12): 240237. doi: 10.12086/oee.2024.240237
Zhang Y, Ma C M, Liu S D, et al. Multi-scale feature enhanced Transformer network for efficient semantic segmentation[J]. Opto-Electron Eng, 2024, 51(12): 240237. doi: 10.12086/oee.2024.240237
[4] Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation[C]//Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015: 234–241. https://doi.org/10.1007/978-3-319-24574-4_28.
[5] Diakogiannis F I, Waldner F, Caccetta P, et al. ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data[J]. ISPRS J Photogramm Remote Sens, 2020, 162: 94−114. doi: 10.1016/j.isprsjprs.2020.01.013
[6] Yin Z J, Liang K M, Ma Z Y, et al. Duplex contextual relation network for polyp segmentation[C]//Proceedings of 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), 2022: 1–5. https://doi.org/10.1109/ISBI52829.2022.9761402.
[7] Lou A G, Guan S Y, Ko H, et al. CaraNet: context axial reverse attention network for segmentation of small medical objects[J]. Proc SPIE, 2022, 12032: 120320D. doi: 10.1117/12.2611802
[8] Huang C H, Wu H Y, Lin Y L. HarDNet-MSEG: a simple encoder-decoder polyp segmentation neural network that achieves over 0.9 mean dice and 86 FPS[Z]. arXiv: 2101.07172, 2021. https://doi.org/10.48550/arXiv.2101.07172.
[9] Shi W T, Xu J, Gao P. SSformer: a lightweight transformer for semantic segmentation[C]//Proceedings of 2022 IEEE 24th International Workshop on Multimedia Signal Processing (MMSP), 2022: 1–5. https://doi.org/10.1109/MMSP55362.2022.9949177.
[10] Wu C, Long C, Li S J, et al. MSRAformer: multiscale spatial reverse attention network for polyp segmentation[J]. Comput Biol Med, 2022, 151: 106274. doi: 10.1016/j.compbiomed.2022.106274
[11] Wang W H, Xie E Z, Li X, et al. PVT v2: improved baselines with pyramid vision transformer[J]. Comput Visual Media, 2022, 8(3): 415−424. doi: 10.1007/s41095-022-0274-8
[12] Ouyang D L, He S, Zhang G Z, et al. Efficient multi-scale attention module with cross-spatial learning[C]//Proceedings of ICASSP 2023–2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2023: 1–5. https://doi.org/10.1109/ICASSP49357.2023.10096516.
[13] Zheng M J, Sun L, Dong J X, et al. SMFANet: a lightweight self-modulation feature aggregation network for efficient image super-resolution[C]//Proceedings of the 18th European Conference on Computer Vision, 2024: 359–375. https://doi.org/10.1007/978-3-031-72973-7_21.
[14] Huo X Z, Sun G, Tian S W, et al. HiFuse: hierarchical multi-scale feature fusion network for medical image classification[J]. Biomed Signal Process Control, 2024, 87: 105534. doi: 10.1016/j.bspc.2023.105534
[15] Chen X K, Lin K Y, Wang J B, et al. Bi-directional cross-modality feature propagation with separation-and-aggregation gate for RGB-D semantic segmentation[C]//Proceedings of the 16th European Conference on Computer Vision, 2020: 561–577. https://doi.org/10.1007/978-3-030-58621-8_33.
[16] Zhang Q L, Yang Y B. SA-Net: shuffle attention for deep convolutional neural networks[C]//Proceedings of ICASSP 2021–2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2021: 2235–2239. https://doi.org/10.1109/ICASSP39728.2021.9414568.
[17] Bernal J, Sánchez F J, Fernández-Esparrach G, et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians[J]. Comput Med Imaging Graphics, 2015, 43: 99−111. doi: 10.1016/j.compmedimag.2015.02.007
[18] Jha D, Smedsrud P H, Riegler M A, et al. Kvasir-SEG: a segmented polyp dataset[C]//Proceedings of the 26th International Conference on MultiMedia Modeling, 2020: 451–462. https://doi.org/10.1007/978-3-030-37734-2_37.
[19] Tajbakhsh N, Gurudu S R, Liang J M. Automated polyp detection in colonoscopy videos using shape and context information[J]. IEEE Trans Med Imaging, 2016, 35(2): 630−644. doi: 10.1109/TMI.2015.2487997
[20] Silva J, Histace A, Romain O, et al. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer[J]. Int J Comput Assisted Radiol Surg, 2014, 9(2): 283−293. doi: 10.1007/s11548-013-0926-3
[21] Dong B, Wang W H, Fan D P, et al. Polyp-PVT: polyp segmentation with pyramid vision transformers[Z]. arXiv: 2108.06932, 2024. https://doi.org/10.48550/arXiv.2108.06932.
[22] 李大湘, 李登辉, 刘颖, 等. 渐进式CNN-Transformer语义补偿息肉分割网络[J]. 光学 精密工程, 2024, 32(16): 2523−2536. doi: 10.37188/OPE.20243216.2523
Li D X, Li D H, Liu Y, et al. Progressive CNN-transformer semantic compensation network for polyp segmentation[J]. Opt Precis Eng, 2024, 32(16): 2523−2536. doi: 10.37188/OPE.20243216.2523
-
访问统计

 E-mail Alert
E-mail Alert RSS
RSS

 下载:
下载: