Colorectal polyp segmentation method combining polarized self-attention and Transformer
-
摘要
针对传统结直肠息肉图像分割方法存在的目标分割不够精确、对比度不足,以及边缘细节模糊等问题,文中结合极化自注意力和Transformer提出了一种新的结直肠息肉图像分割方法。首先,设计了一种改进的相位感知混合模块,通过动态捕捉Transformer结直肠息肉图像的多尺度上下文信息,以使目标分割更加精确。其次,在新方法中引入了极化自注意力机制,实现了图像的自我注意力强化,使得到的图像特征可以直接用于息肉分割任务中,以达到提高病灶区域与正常组织区域对比度的目的。另外,利用线索交叉融合模块加强动态分割时对图像几何结构的捕捉能力,以达到提升结果图像边缘细节的目的。实验结果表明,文中提出的方法不仅能够有效地提升结直肠息肉分割的精确度和对比度,并且还能够较好地克服分割图像细节模糊的问题。在数据集CVC-ClinicDB、Kvasir 、CVC-ColonDB和ETIS-LaribPolypDB上的测试结果表明,文中所提新方法能够取得更好的分割效果,其Dice相似性指数分别为0.946、0.927、0.805和0.781。
-
关键词:
- 结直肠息肉 /
- Transformer /
- 相位感知模块 /
- 极化自注意力模块
Abstract
A new colorectal polyp image segmentation method combining polarizing self-attention and Transformer is proposed to solve the problems of traditional colorectal polyp image segmentation such as insufficient target segmentation, insufficient contrast and blurred edge details. Firstly, an improved phase sensing hybrid module is designed to dynamically capture multi-scale context information of colorectal polyp images in Transformer to make target segmentation more accurate. Secondly, the polarization self-attention mechanism is introduced into the new method to realize the self-attention enhancement of the image, so that the obtained image features can be directly used in the polyp segmentation task to improve the contrast between the lesion area and the normal tissue area. In addition, the cue-cross fusion module is used to enhance the ability to capture the geometric structure of the image in dynamic segmentation, so as to improve the edge details of the resulting image. The experimental results show that the proposed method can not only effectively improve the precision and contrast of colorectal polyp segmentation, but also overcome the problem of blurred detail in the segmentation image. The test results on the data sets CVC-ClinicDB, Kvasir, CVC-ColonDB and ETIS-LaribPolypDB show that the proposed method can achieve better segmentation results, and the Dice similarity index is 0.946, 0.927, 0.805 and 0.781, respectively.
-
Key words:
- colorectal polyp /
- Transformer /
- phase sensing module /
- polarized self-attention module
-
Overview
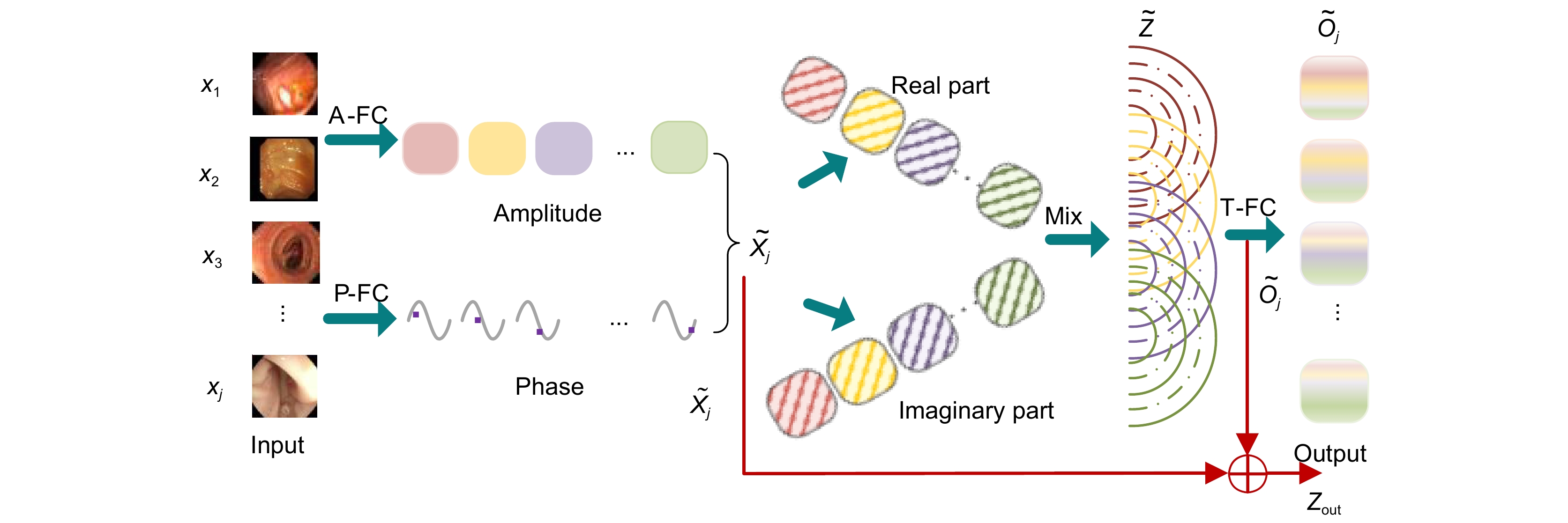
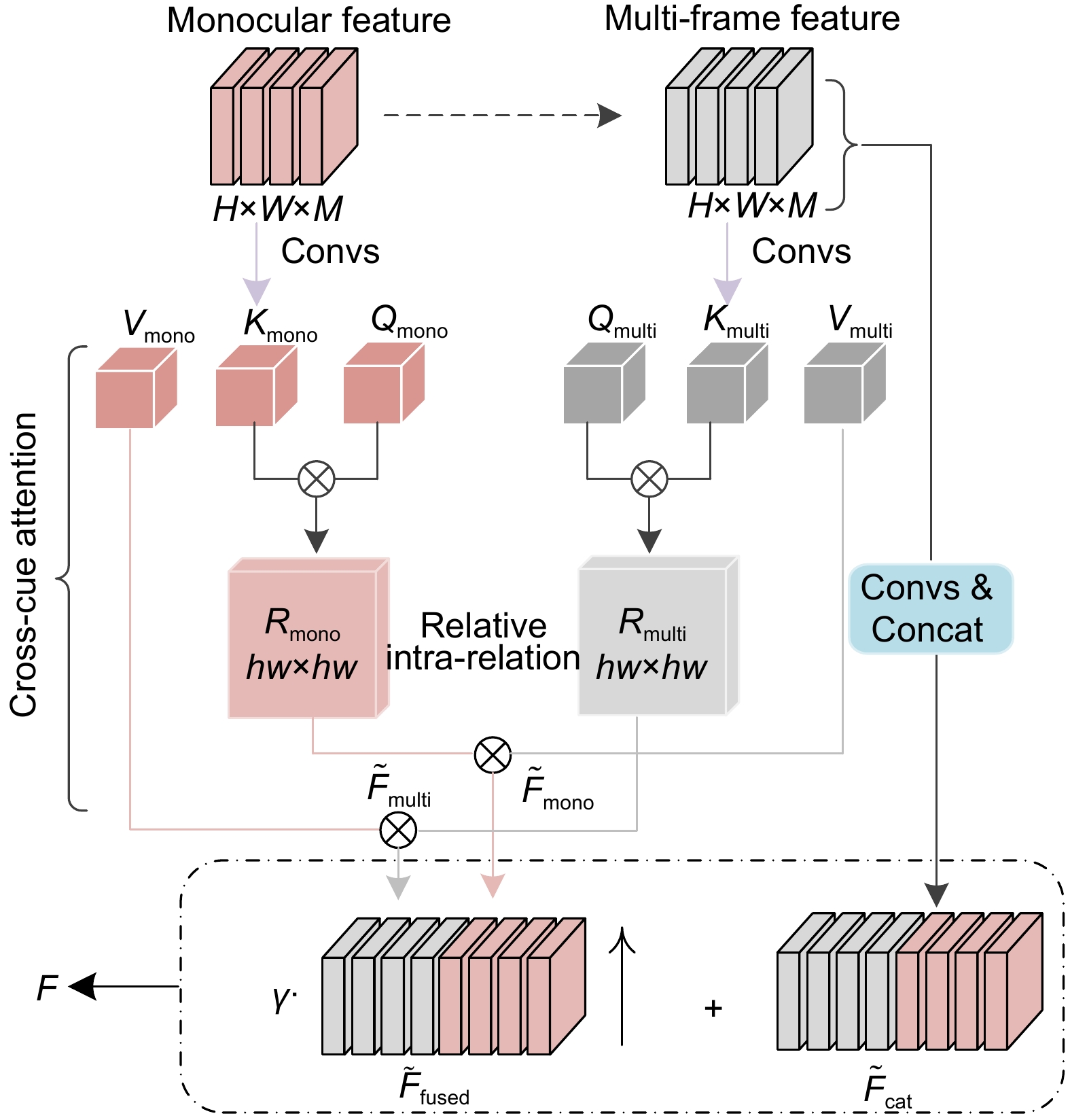
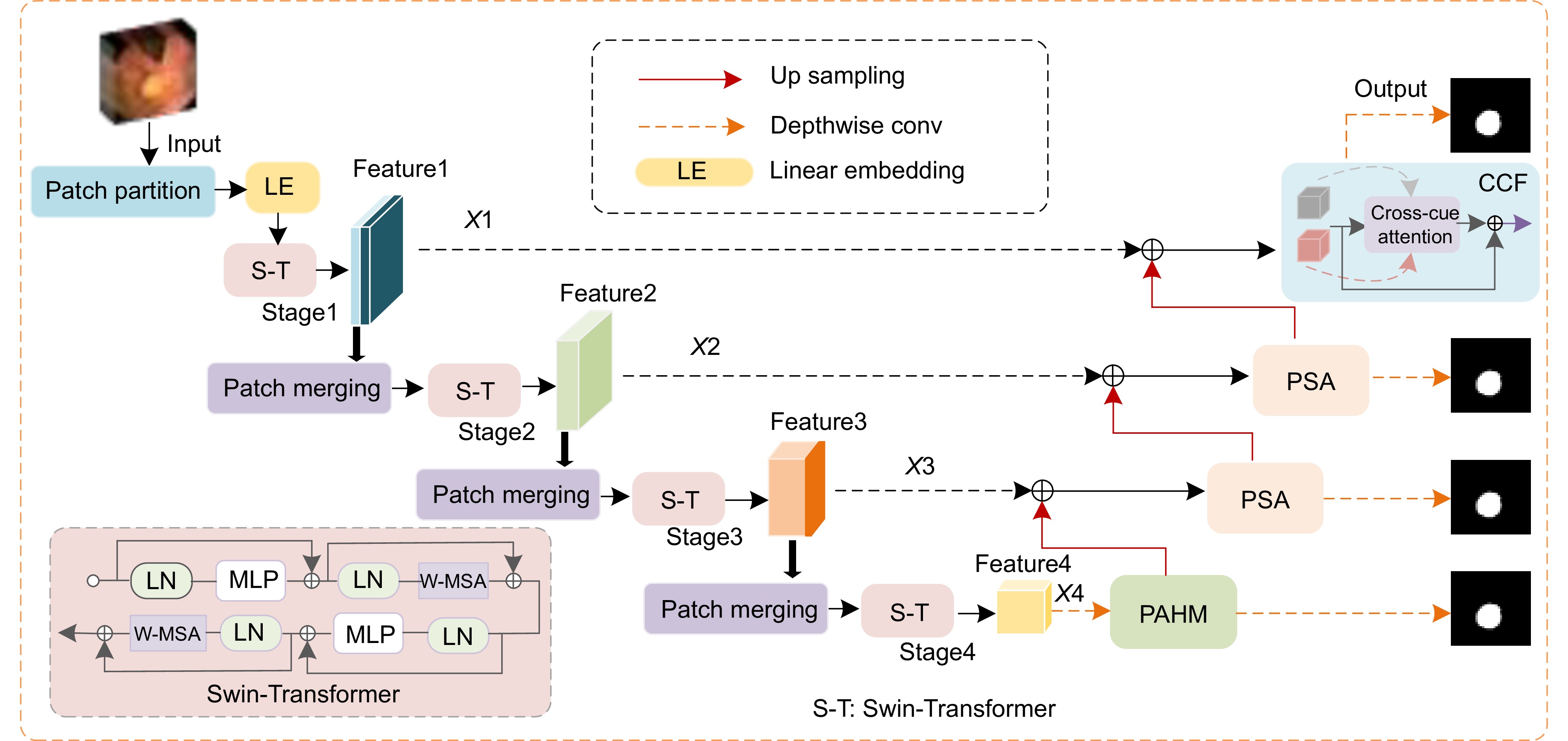
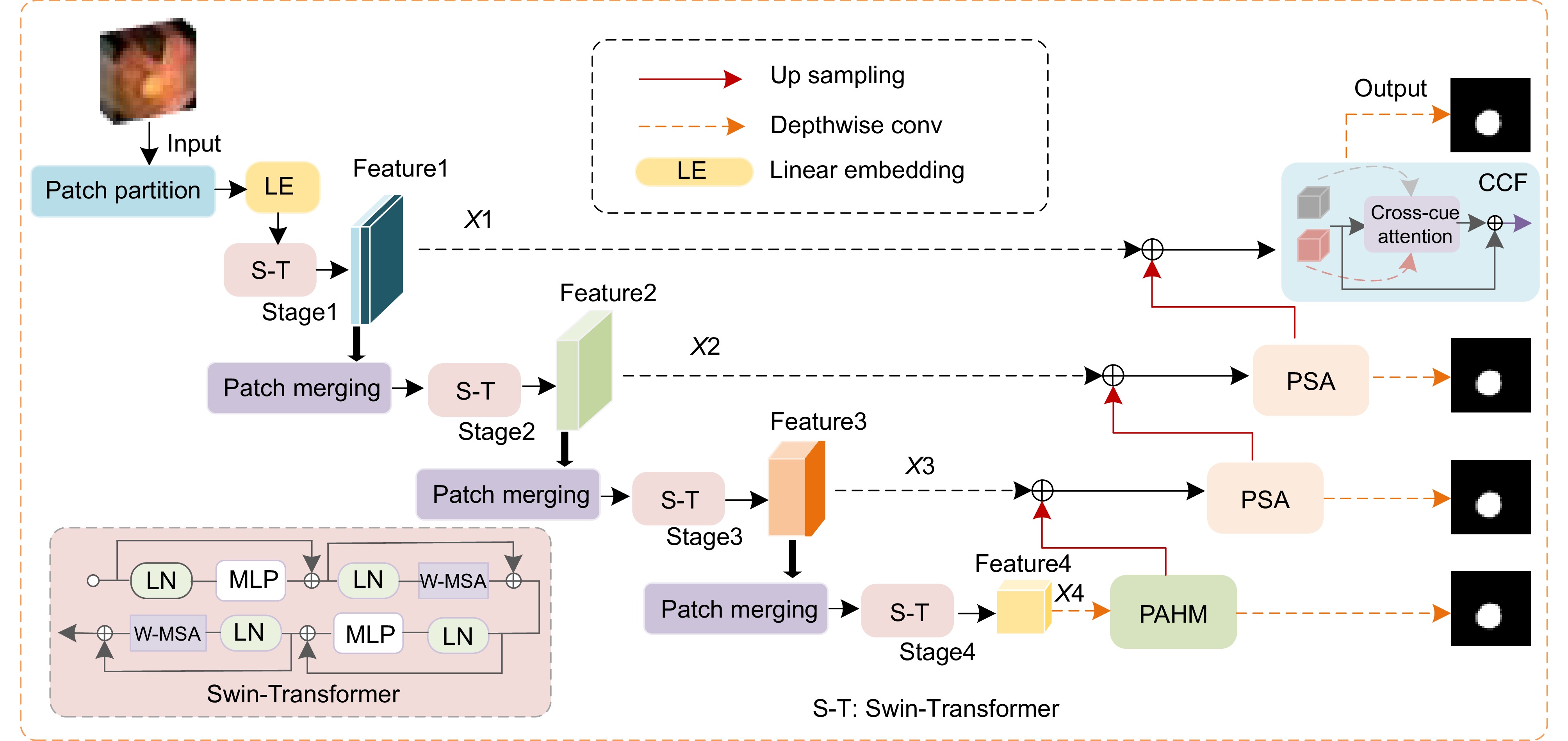
Overview: Among malignant diseases, colorectal cancer is one of the most common cancers in life, and its morbidity and mortality have been high. Therefore, it is urgent to develop an automatic recognition and automatic segmentation algorithm for colorectal polyp image segmentation to help doctors improve the efficiency of diagnosing patients. However, the traditional colorectal polyp segmentation method requires manual extraction of lesion features and the integration strategy will over-rely on the experience of the implementor. Therefore, the traditional colorectal polyp segmentation method is prone to problems such as inaccurate target segmentation, insufficient contrast and blurred edge details during segmentation. In order to solve the problems existing in the traditional method, In this paper, a new colorectal polyp segmentation network TPSA-Net, which combines polarized self-attention and Transformer, is proposed. Firstly, in order to make better use of the semantic information of image blocks at different phase levels to improve the segmentation accuracy of target images, an improved phase sensing hybrid module is designed in this paper, which can dynamically capture multi-scale context information at different levels of colorectal polyp images to improve the accuracy of target segmentation. Secondly, the polarization self-attention module is introduced to fully consider the characteristics of pixels and strengthen the self-attention of the image, so as to improve the contrast between the lesion area and the normal tissue area. Finally, the dynamic capturing ability of the geometric structure of the image was enhanced by the cross-fusion module of the clues, and the complementary characteristics of the two clues in single/multi-frame were improved to solve the problem of blurred edge details during colorectal polyp segmentation. Experiments were conducted on four datasets, CVC-ClinicDB, Kvasir, CVC-ColonDB and ETIS-LaribPolypDB, and the Dice similarity index was 0.946, 0.927, 0.805 and 0.781, respectively. Compared with U-Net, the traditional medical image segmentation network was improved by 12.4%, 14.5%, 29.3% and 37.5 respectively. The average MIou intersection ratio index was 0.901, 0.880, 0.729 and 0.706, respectively, which had certain application value in the diagnosis of colorectal polyps. A large number of experimental results show that the TPSA-Net method proposed in this paper can not only effectively improve the accuracy and contrast of colorectal polyp segmentation, but also overcome the problem of blurred detail in the segmentation image. How to use deep learning technology to research more simple and efficient colorectal polyp segmentation methods is the future focus.
-
-
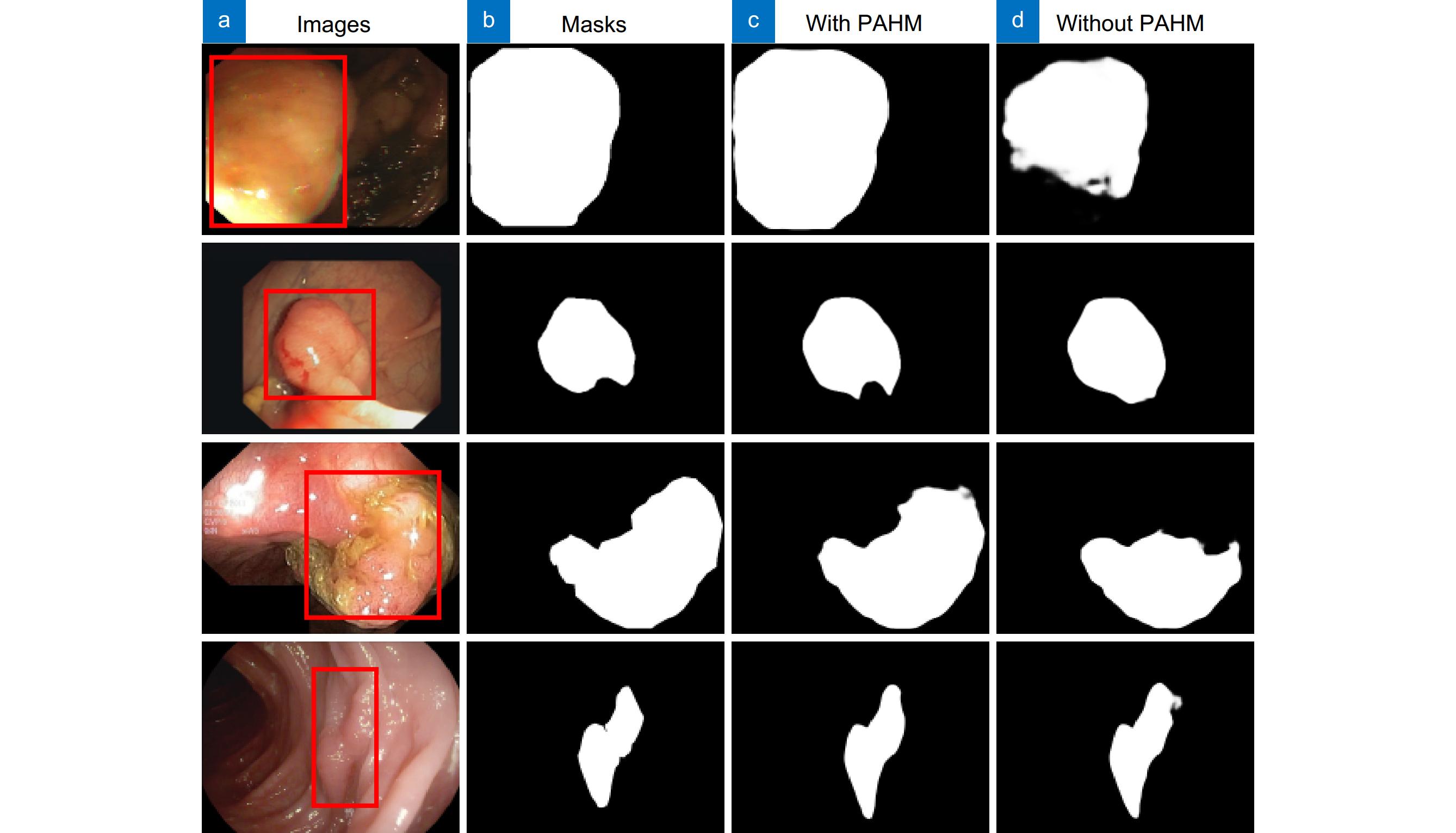
表 1 有/无PAHM在CVC-ClinicDB和CVC-ColonDB上的对比
Table 1. Comparison with/without PAHM on CVC-ClinicDB and CVC-ColonDB
Dataset Method Dice MIoU SE CVC-ClinicDB N1 0.942 0.898 0.950 N4 0.946 0.901 0.951 CVC-ColonDB N1 0.800 0.727 0.819 N4 0.805 0.729 0.822 表 2 有/无PSA在CVC-ClinicDB和CVC-ColonDB上的对比
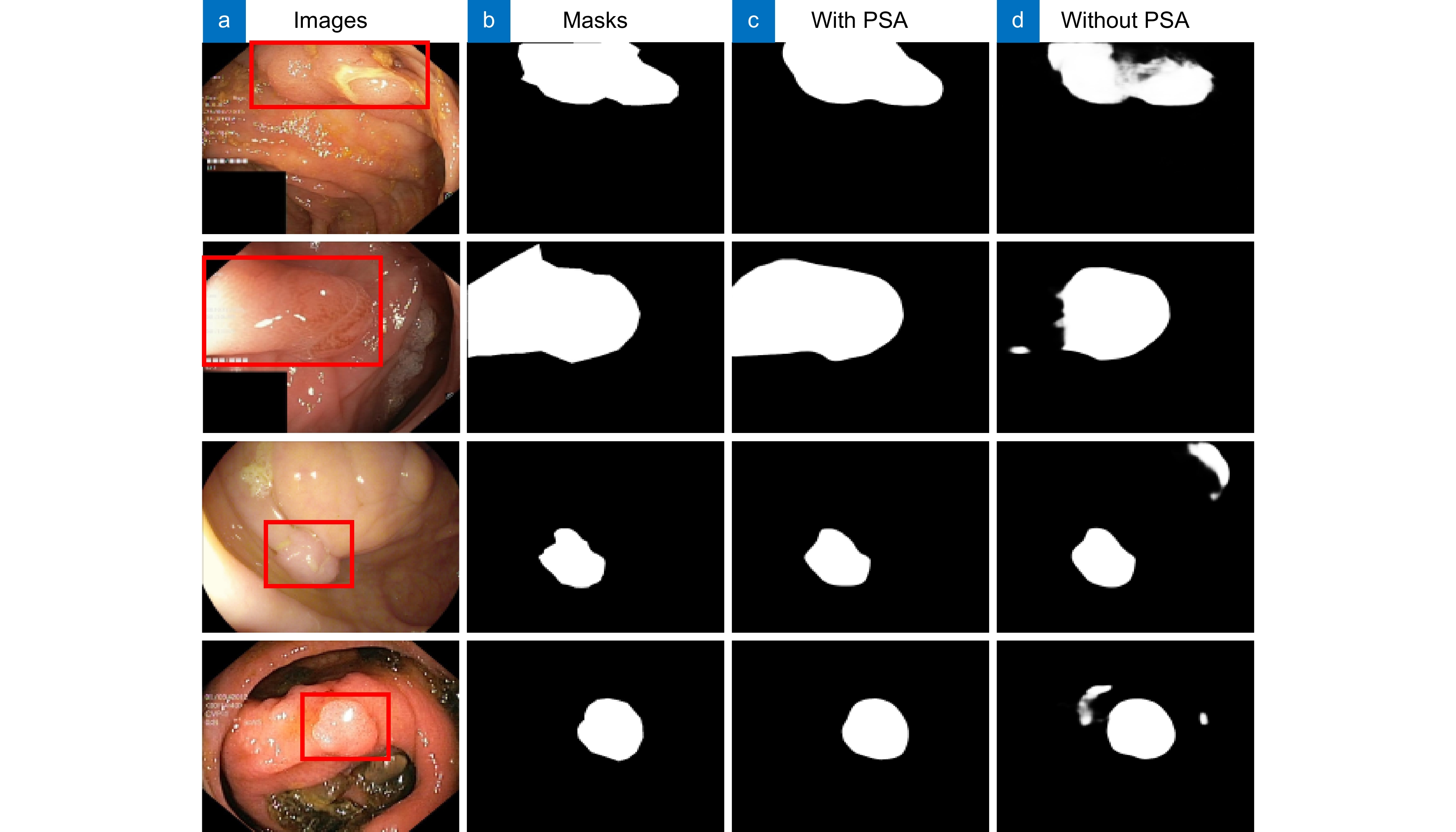
Table 2. Comparison with/without PSA on CVC-ClinicDB and CVC-ColonDB
Dataset Method Dice MIoU SE CVC-ClinicDB N2 0.937 0.881 0.946 N4 0.946 0.901 0.951 CVC-ColonDB N2 0.788 0.711 0.813 N4 0.805 0.729 0.822 表 3 有/无CCF在CVC-ClinicDB和CVC-ColonDB上的对比
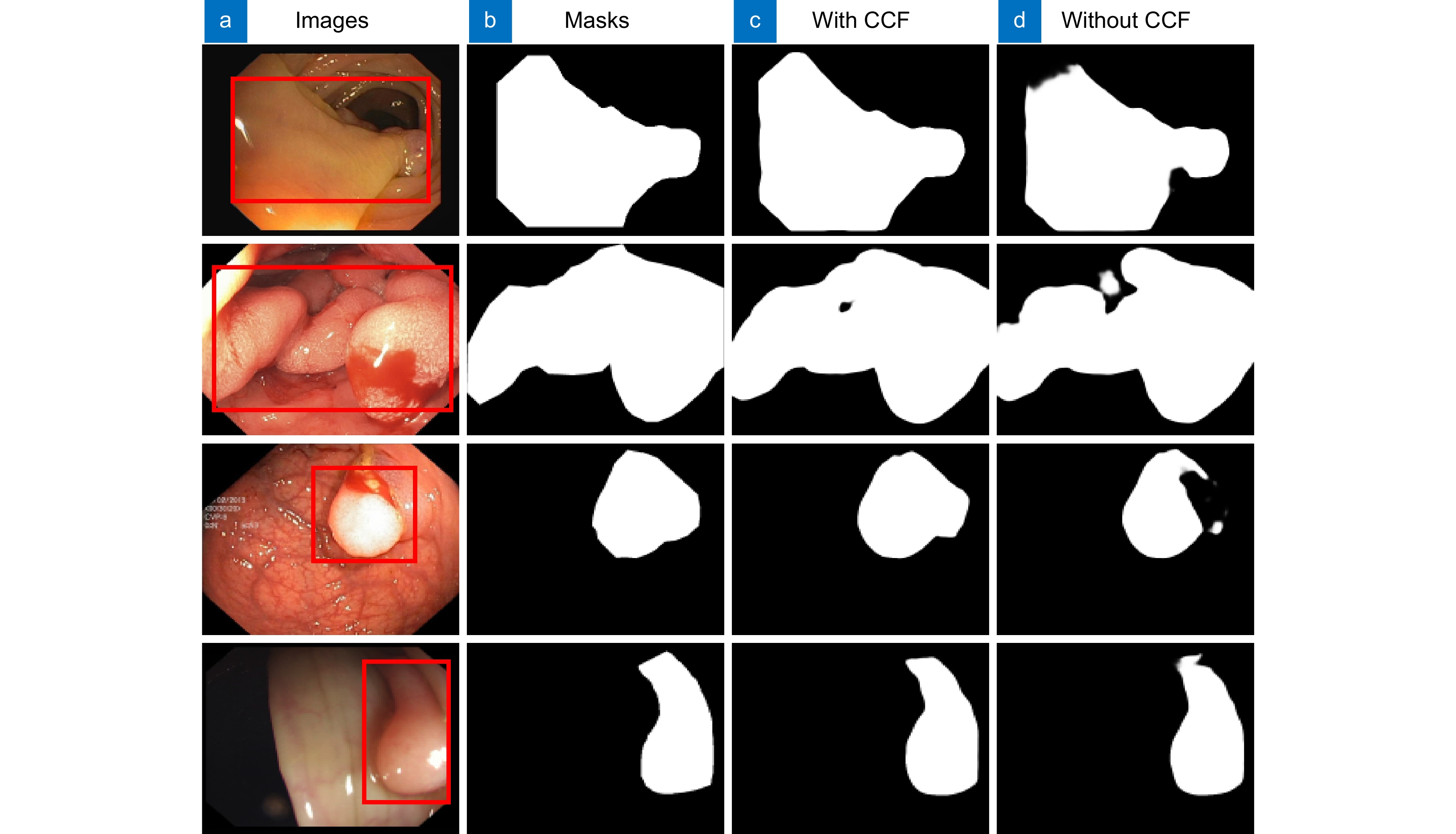
Table 3. Comparison with/without CCF on CVC-ClinicDB and CVC-ColonDB
Dataset Method Dice MIoU SE CVC-ClinicDB N3 0.942 0.894 0.949 N4 0.946 0.901 0.951 CVC-ColonDB N3 0.751 0.684 0.777 N4 0.805 0.729 0.822 表 4 实验参数设置
Table 4. Experimental parameter settings
Dataset Traindata Testdata Picture size/pixel CVC-ClinicDB 550 62 352×352 Kvasir 900 100 352×352 ETIS-LaribPolypDB 0 196 352×352 CVC-ColonDB 0 380 352×352 表 5 不同算法在CVC-ClinicDB和Kvasir上的对比
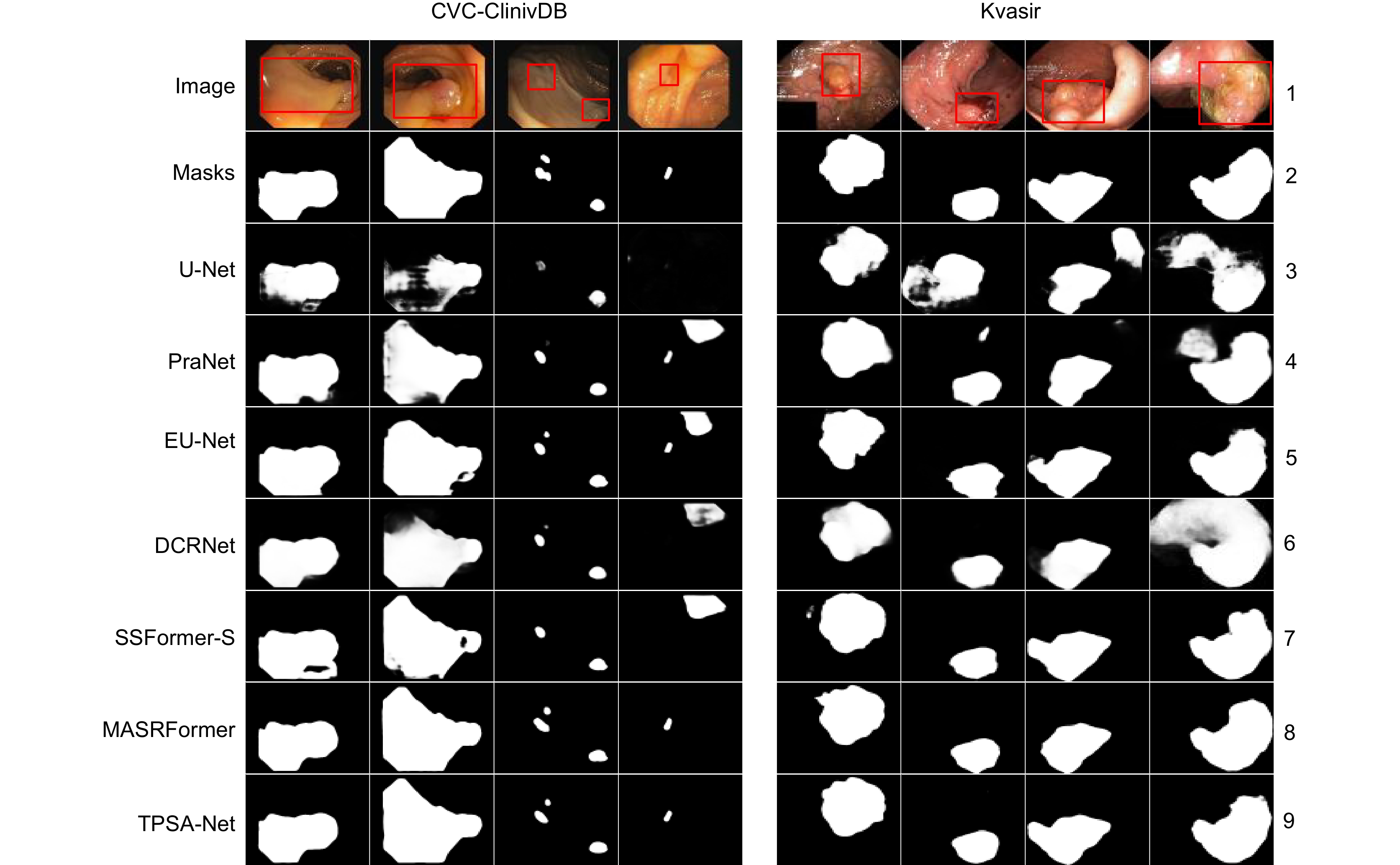
Table 5. Comparison of different algorithms on CVC-ClinicDB and Kvasir
Dataset Method Dice MIoU SE PC F2 MAE CVC-
ClinicDBU-Net 0.822 0.756 0.836 0.835 0.828 0.020 PraNet 0.902 0.850 0.911 0.905 0.901 0.009 EU-Net 0.905 0.849 0.956 0.881 0.927 0.011 DCRNet 0.899 0.847 0.912 0.893 0.907 0.010 SSFormer-S 0.919 0.872 0.903 0.939 0.908 0.007 MSRAFormer 0.934 0.884 0.950 0.924 0.944 0.007 Ours 0.946 0.901 0.957 0.943 0.949 0.005 Kvasir U-Net 0.821 0.747 0.855 0.856 0.828 0.055 PraNet 0.901 0.841 0.910 0.916 0.903 0.030 EU-Net 0.911 0.858 0.931 0.912 0.919 0.028 DCRNet 0.889 0.823 0.903 0.902 0.892 0.034 SSFormer-S 0.925 0.876 0.917 0.944 0.921 0.020 MSRAFormer 0.919 0.870 0.921 0.938 0.918 0.020 Ours 0.927 0.880 0.932 0.950 0.923 0.020 表 6 不同算法在CVC-ColonDB和ETIS-LaribPolypDB上的对比
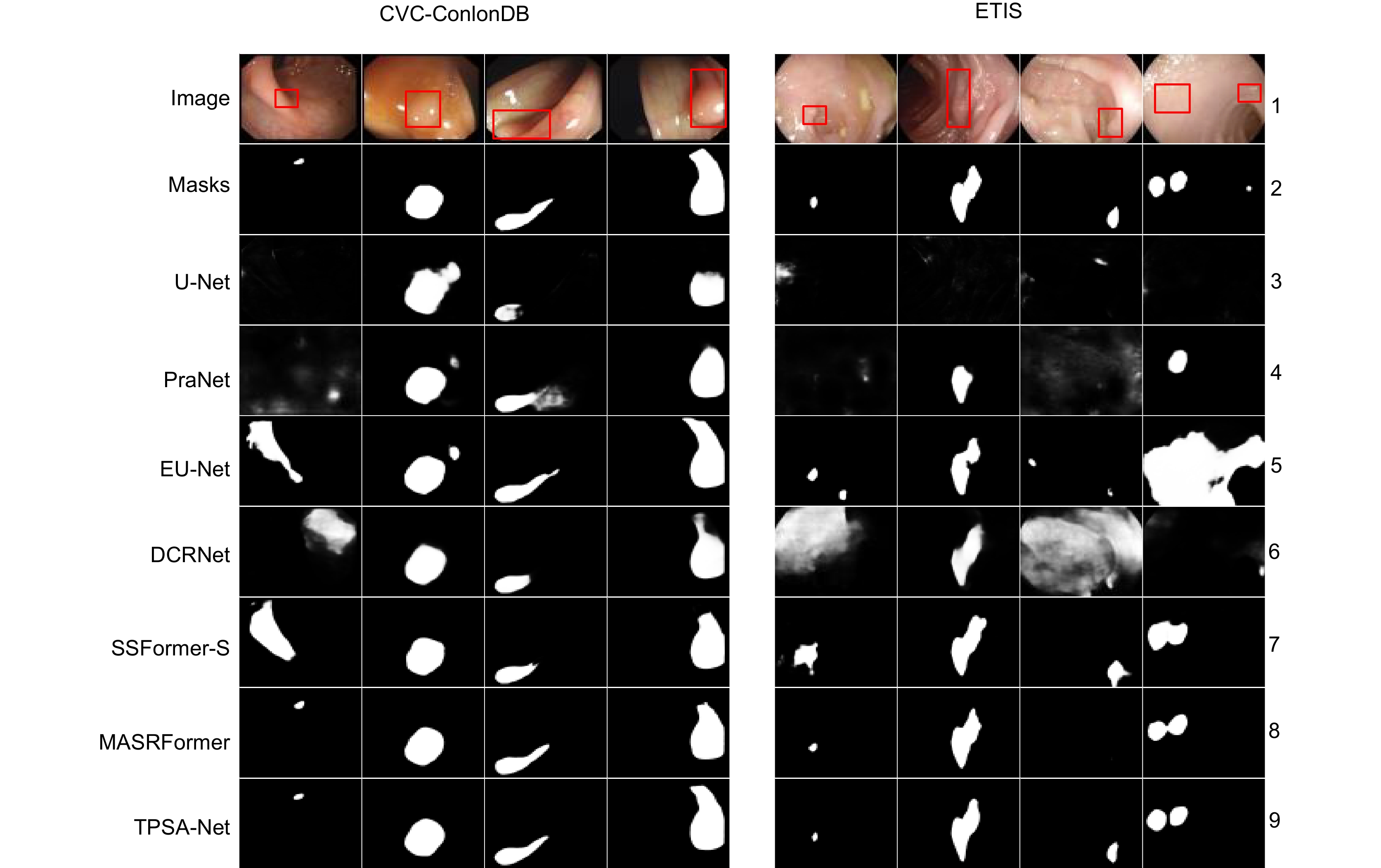
Table 6. Comparison of different algorithms on CVC-ColonDB and ETIS-LaribPolypDB
Dataset Method Dice MIoU SE PC F2 MAE CVC-
ColonDBU-Net 0.512 0.438 0.524 0.621 0.510 0.059 PraNet 0.717 0.641 0.740 0.755 0.716 0.044 EU-Net 0.756 0.683 0.848 0.756 0.789 0.043 DCRNet 0.707 0.632 0.777 0.719 0.723 0.051 SSFormer-S 0.775 0.698 0.776 0.836 0.767 0.034 MSRAFormer 0.765 0.695 0.801 0.870 0.772 0.031 Ours 0.805 0.729 0.878 0.872 0.806 0.025 ETIS-
LaribPolypDBU-Net 0.406 0.334 0.482 0.439 0.428 0.037 PraNet 0.631 0.567 0.689 0.628 0.649 0.030 EU-Net 0.690 0.611 0.871 0.637 0.749 0.065 DCRNet 0.548 0.484 0.744 0.504 0.600 0.095 SSFormer-S 0.770 0.695 0.856 0.744 0.782 0.017 MSRAFormer 0.749 0.674 0.821 0.787 0.782 0.012 Ours 0.781 0.706 0.874 0.808 0.807 0.011 表 7 各模块在Kvasir和EITS数据集上的消融研究
Table 7. Ablation of each module on Kvasir and EITS datasets
Method CCF PAHM PSA Kvasir ETIS Dice MIoU SE F2 Dice MIoU SE F2 M1 × √ √ 0.919 0.873 0.919 0.920 0.744 0.674 0.803 0.769 M2 √ × √ 0.918 0.872 0.913 0.914 0.740 0.672 0.802 0.767 M3 √ √ × 0.924 0.876 0.924 0.918 0.756 0.681 0.836 0.792 M4 √ √ √ 0.927 0.880 0.926 0.923 0.781 0.706 0.874 0.807 -
参考文献
[1] Liang H, Cheng Z M, Zhong H Q, et al. A region-based convolutional network for nuclei detection and segmentation in microscopy images[J]. Biomed Signal Process Control, 2022, 71: 103276. doi: 10.1016/j.bspc.2021.103276
[2] Jha D, Smedsrud P H, Johansen D, et al. A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation[J]. IEEE J Biomed Health Inform, 2021, 25(6): 2029−2040. doi: 10.1109/JBHI.2021.3049304
[3] Li W S, Zhao Y H, Li F Y, et al. MIA-Net: multi-information aggregation network combining transformers and convolutional feature learning for polyp segmentation[J]. Knowl-Based Syst, 2022, 247: 108824. doi: 10.1016/j.knosys.2022.108824
[4] 丁俊华, 袁明辉. 基于双分支多尺度融合网络的毫米波SAR图像多目标语义分割方法[J]. 光电工程, 2023, 50(12): 230242. doi: 10.12086/oee.2023.230242
Ding J H, Yuan M H. A multi-target semantic segmentation method for millimetre wave SAR images based on a dual-branch multi-scale fusion network[J]. Opto-Electron Eng, 2023, 50(12): 230242. doi: 10.12086/oee.2023.230242
[5] Vala M H J, Baxi A. A review on Otsu image segmentation algorithm[J]. Int J Adv Res Comput Eng Technol, 2013, 2(2): 387−389.
[6] Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations[J]. IEEE Trans Pattern Anal Mach Intell, 1991, 13(6): 583−598. doi: 10.1109/34.87344
[7] Canny J. A computational approach to edge detection[J]. IEEE Trans Pattern Anal Mach Intell, 1986, PAMI-8(6): 679−698. doi: 10.1109/TPAMI.1986.4767851
[8] Liang Y B, Fu J. Watershed algorithm for medical image segmentation based on morphology and total variation model[J]. Int J Patt Recogn Artif Intell, 2019, 33(5): 1954019. doi: 10.1142/S0218001419540193
[9] Ali S M F, Khan M T, Haider S U, et al. Depth-wise separable atrous convolution for polyps segmentation in gastro-intestinal tract[C]//Proceedings of the Working Notes Proceedings of the MediaEval 2020 Workshop, 2021.
[10] Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: transformers for image recognition at scale[C]//Proceedings of the 9th International Conference on Learning Representations, 2021.
[11] Wang W H, Xie E Z, Li X, et al. Pyramid vision transformer: a versatile backbone for dense prediction without convolutions[C]//Proceedings of 2021 IEEE/CVF International Conference on Computer Vision, 2021: 548–558. https://doi.org/10.1109/ICCV48922.2021.00061.
[12] Wu C, Long C, Li S J, et al. MSRAformer: multiscale spatial reverse attention network for polyp segmentation[J]. Comput Biol Med, 2022, 151: 106274. doi: 10.1016/j.compbiomed.2022.106274
[13] Liu Z, Lin Y T, Cao Y, et al. Swin transformer: hierarchical vision transformer using shifted windows[C]//Proceedings of 2021 IEEE/CVF International Conference on Computer Vision, 2021: 9992–10002. https://doi.org/10.1109/ICCV48922.2021.00986.
[14] Tang Y H, Han K, Guo J Y, et al. An image patch is a wave: phase-aware vision MLP[C]//Proceedings of 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2022: 10925–10934. https://doi.org/10.1109/CVPR52688.2022.01066.
[15] Liu H J, Liu F Q, Fan X Y, et al. Polarized self-attention: towards high-quality pixel-wise regression[Z]. arXiv: 2107.00782, 2021. https://arxiv.org/abs/2107.00782.
[16] Li R, Gong D, Yin W, et al. Learning to fuse monocular and multi-view cues for multi-frame depth estimation in dynamic scenes[C]//Proceedings of 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2023: 21539–21548. https://doi.org/10.1109/CVPR52729.2023.02063.
[17] Bernal J, Sánchez F J, Fernández-Esparrach G, et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians[J]. Comput Med Imaging Graph, 2015, 43: 99−111. doi: 10.1016/j.compmedimag.2015.02.007
[18] Amsaleg L, Huet B, Larson M, et al. Proceedings of the 27th ACM international conference on multimedia[C]. New York: ACM Press, 2019.
[19] Silva J, Histace A, Romain O, et al. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer[J]. Int J Comput Assist Radiol Surg, 2014, 9(2): 283−293. doi: 10.1007/s11548-013-0926-3
[20] Tajbakhsh N, Gurudu S R, Liang J M. Automated polyp detection in colonoscopy videos using shape and context information[J]. IEEE Trans Med Imaging, 2016, 35(2): 630−644. doi: 10.1109/TMI.2015.2487997
[21] Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation[C]//Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015: 234–241. https://doi.org/10.1007/978-3-319-24574-4_28.
[22] Fan D P, Ji G P, Zhou T, et al. PraNet: parallel reverse attention network for polyp segmentation[C]//Proceedings of the 23rd International Conference on Medical Image Computing and Computer-Assisted Intervention, 2020: 263–273. https://doi.org/10.1007/978-3-030-59725-2_26.
[23] Patel K, Bur A M, Wang G H. Enhanced U-Net: a feature enhancement network for polyp segmentation[C]//Proceedings of 2021 18th Conference on Robots and Vision, 2021: 181–188. https://doi.org/10.1109/CRV52889.2021.00032.
[24] Yin Z J, Liang K M, Ma Z Y, et al. Duplex contextual relation network for polyp segmentation[C]//Proceedings of 2022 IEEE 19th International Symposium on Biomedical Imaging, 2022: 1–5. https://doi.org/10.1109/ISBI52829.2022.9761402.
[25] Wang J F, Huang Q M, Tang F L, et al. Stepwise feature fusion: local guides global[C]//Proceedings of the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention, 2022: 110–120. https://doi.org/10.1007/978-3-031-16437-8_11.
-
访问统计

 E-mail Alert
E-mail Alert RSS
RSS
 下载:
下载: