-
摘要
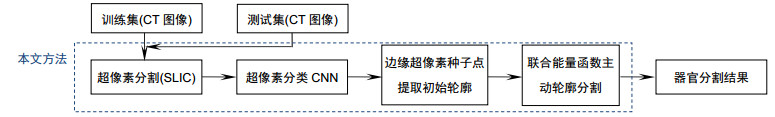
为解决医学CT图像主动轮廓分割方法中对初始轮廓敏感的问题,提出一种基于超像素和卷积神经网络的人体器官CT图像联合能量函数主动轮廓分割方法。该方法首先基于超像素分割对CT图像进行超像素网格化,并通过卷积神经网络进行超像素分类确定边缘超像素;然后提取边缘超像素的种子点组成初始轮廓;最后在提取的初始轮廓基础上,通过求解本文提出的综合能量函数最小值实现人体器官分割。实验结果表明,本文方法与先进的U-Net方法相比平均Dice系数提高5%,为临床CT图像病变诊断提供理论基础和新的解决方案。

Abstract
In this paper, an active contour segmentation method for organs CT images based on super-pixel and convolutional neural network is proposed to solve the sensitive problem of the initial contour of the segmentation method of the CT image. The method firstly super-pixels the CT image based on super-pixel segmentation and determines the edge super-pixels by the super-pixel classification through a convolutional neural network. Afterwards, the seed points of the edge super-pixels are extracted to form the initial contour. Finally, based on the extracted initial contour, the human organ segmentation is realized by solving the minimum value of the integrated energy function proposed in this paper. The results in this paper show that the average Dice coefficient is improved by 5% compared with the advanced U-Net method, providing a theoretical basis and a new solution for the diagnosis of clinical CT image lesions.
-
Overview

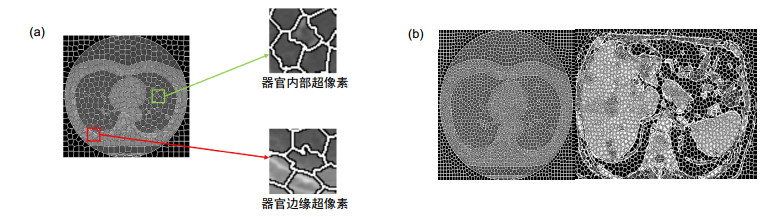
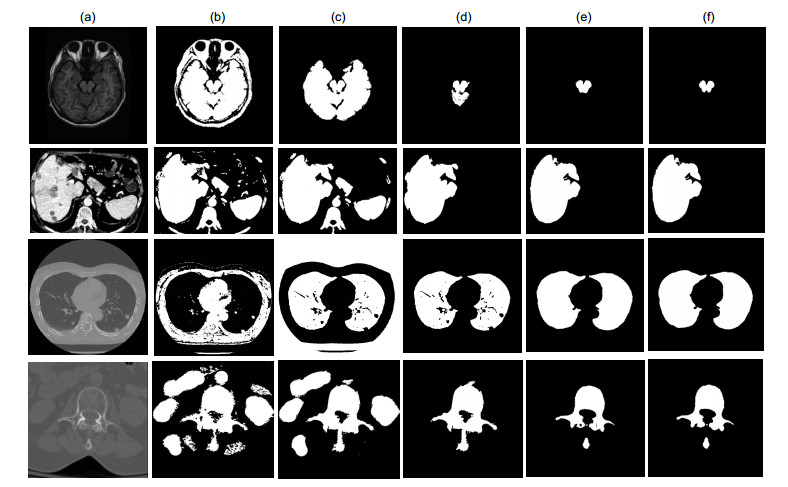
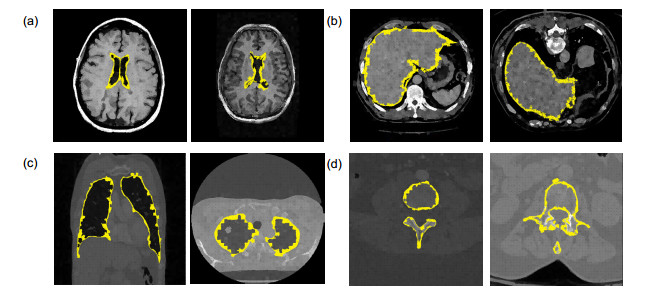
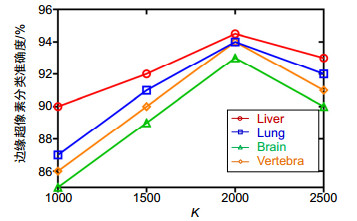
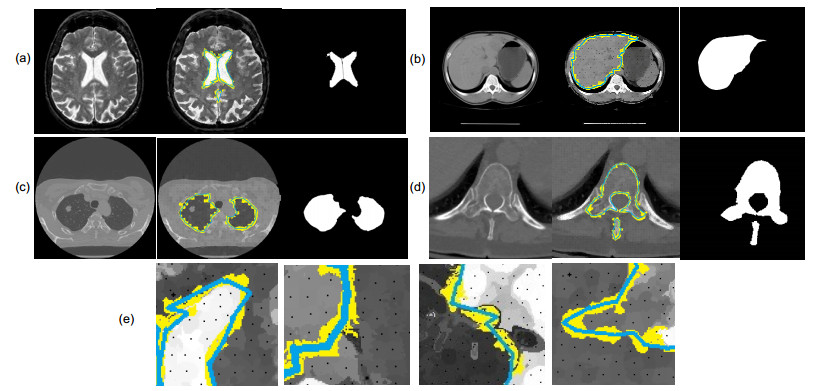
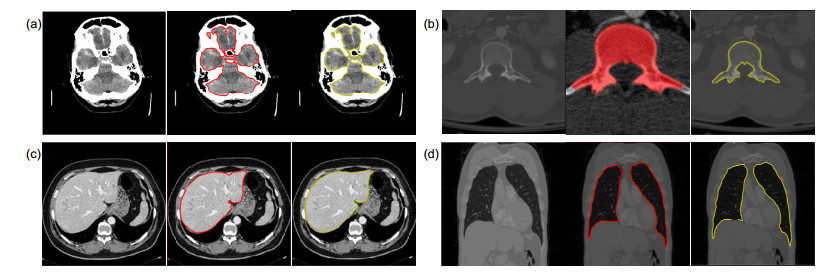
Overview: Computed tomography images have the advantage of fast imaging speed and sharp imaging. CT images are one of the most important medical imaging techniques for human evaluation and it has become a conventional means of daily inspection. For computer-aided diagnosis, interest towards segmentation of regions in CT images is an essential prerequisite. Therefore, it is imperative to seek an automatic CT image method that can replace manual segmentation. This paper presents a fully automated CT image segmentation method for human organs. Firstly, super-pixel meshing is performed on CT images based on the super-pixel segmentation, and super-pixel classification is performed by a convolutional neural network to determine edge super-pixels. Then, seed points of edge super-pixels are extracted to form initial contours. Finally, the initial contour is obtained based on the extraction by solving the minimum of the integrated energy function proposed herein. In order to comprehensively evaluate the segmentation effect of this method on medical CT images, this paper mainly divides CT image experiments into four organs, including the brain, liver, lungs, and vertebral body. The experimental results show that the super-pixel classification CNN has achieved excellent results in the super-pixel classification of CT images. The classification accuracy reaches 92%. The initial contour of the super-pixel seed points extracted in this paper is close to the organ edge, and the next contour based on a significant amount of time is stored in the solution of the integrated energy function. For the target image segmentation of brain, liver, lung, and vertebrae, the proposed method can accurately locate the edge super-pixels that completely extract the initial contour of the edge super-pixel seed point structure, and complete the segmentation contour subdivision by minimizing the improved integrated energy function. Compared with the advanced U-net method, the average Dice coefficient of the proposed method increase by 5%. It may provide a theoretical basis and a new solution for the diagnosis of clinical CT image lesions. In general, this approach can reduce time and improve efficiency while ensuring segmentation accuracy. In the future study, efforts would be made to test the framework on other types of medical images, such as MRI images and ultrasound images. At the same time, we also look forward to improving accuracy and efficiency and incorporating this framework into clinical diagnostics that benefit patients.
-
-
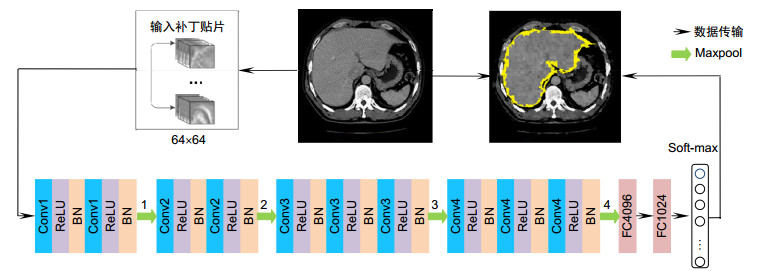
表 1 超像素分类CNN网络结构参数
Table 1. The parameters of super-pixel classification CNN network
Layer Kernel Stride Pad Output Data - - - 64×64 Conv1 BN 2×2 - 1 0 64×64 - 0 64×64 Maxpool1 2×2 2 0 32×32 Conv2 2×2 1 0 32×32 Maxpool2 2×2 2 0 16×16 Conv3 2×2 1 0 16×16 Maxpool3 2×2 2 1 8×8 Conv4 2×2 1 0 8×8 Maxpool4 2×2 1 0 4×4 FC1 FC-4096 FC2 FC-1024 Soft-max Soft-max lables=1、0 表 2 数据表
Table 2. Data sheet
器官 数据集 脑 BrainWeb:脑数据库数据集,20组模型 椎骨 SpineWeb:腰椎分割CT图像数据库,3000张512×512切片 肝脏 Segmentation of the Liver:肝脏分割数据集,5500张512×512切片 肺 TIANCHI:开放的中文数据集,5000张512×512切片 表 3 定量实验分割结果
Table 3. Quantitative experiment segmentation results
指标 任意初始轮廓 定位初始轮廓 本文方法 无边缘能量 有边缘能量 无边缘能量 有边缘能量 初始轮廓+边缘能量+后处理 Jaccard 0.545 0.773 0.881 0.943 0.945 Dice 0.548 0.778 0.892 0.947 0.948 CCR 0.547 0.775 0.890 0.944 0.945 分割耗时/(s/片) 45 43 23 27 25 表 4 分割结果各评价指标
Table 4. Evaluation indicators of segmentation results
器官 超像素分类准确度 Jaccard Dice CCR 脑 0.928 0.955 0.955 0.954 椎骨 0.931 0.941 0.949 0.943 肝脏 0.940 0.943 0.976 0.977 0.976 肺 0.943 0.975 0.978 0.977 -
参考文献
[1] Moltz J H, Bornemann L, Dicken V, et al. Segmentation of liver metastases in CT scans by adaptive thresholding and morphological processing[C]//The MIDAS Journal-Grand Challenge Liver Tumor Segmentation (2008 MICCAI Workshop), 2008, 472: 195-222.
[2] Chang Y L, Li X B. Adaptive image region-growing[J]. IEEE Transactions on Image Processing, 1994, 3(6): 868-872. doi: 10.1109/83.336259
[3] Pohle R, Toennies K D. Segmentation of medical images using adaptive region growing[J]. Proceedings of SPIE, 2001, 4322: 1337-1346. doi: 10.1117/12.431013
[4] Oda M, Nakaoka T, Kitasaka T, et al. Organ segmentation from 3D abdominal CT images based on atlas selection and graph cut[C]//Proceedings of the Third International Conference on Abdominal Imaging: Computational and Clinical Applications, 2012, 7029: 181-188.
[5] Criminisi A, Shotton J, Robertson D, et al. Regression forests for efficient anatomy detection and localization in CT studies[C]//International MICCAI Workshop, MCV 2010, 2011: 106-117.
[6] 唐利明, 田学全, 黄大荣, 等.结合FCMS与变分水平集的图像分割模型[J].自动化学报, 2014, 40(6): 1233-1248. http://d.old.wanfangdata.com.cn/Periodical/zdhxb201406022
Tang L M, Tian X Q, Huang D R, et al. Image segmentation model combined with FCMS and variational level set[J]. Acta Automatica Sinica, 2014, 40(6): 1233-1248. http://d.old.wanfangdata.com.cn/Periodical/zdhxb201406022
[7] 陈侃, 李彬, 田联房.基于模糊速度函数的活动轮廓模型的肺结节分割[J].自动化学报, 2013, 39(8): 1257-1264. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=zdhxb201308010
Chen K, Li B, Tian L F. A segmentation algorithm of pulmonary nodules using active contour model based on fuzzy speed function[J]. Acta Automatica Sinica, 2013, 39(8): 1257-1264. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=zdhxb201308010
[8] 孙文燕, 董恩清, 曹祝楼, 等.一种基于模糊主动轮廓的鲁棒局部分割方法[J].自动化学报, 2017, 43(4): 611-621. http://d.old.wanfangdata.com.cn/Periodical/zdhxb201704011
Sun W Y, Dong E Q, Cao Z L, et al. A robust local segmentation method based on fuzzy-energy based active contour[J]. Acta Automatica Sinica, 2017, 43(4): 611-621. http://d.old.wanfangdata.com.cn/Periodical/zdhxb201704011
[9] Jones J L, Xie X H, Essa E. Combining region-based and imprecise boundary-based cues for interactive medical image segmentation[J]. International Journal for Numerical Methods in Biomedical Engineering, 2014, 30(12): 1649-1666. doi: 10.1002/cnm.2693
[10] Tah A A, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool[J]. BMC Medical Imaging, 2015, 15(1): 29. doi: 10.1186/s12880-015-0068-x
[11] Criminisi A, Robertson D, Konukoglu E, et al. Regression forests for efficient anatomy detection and localization in computed tomography scans[J]. Medical Image Analysis, 2013, 17(8): 1293-1303. doi: 10.1016/j.media.2013.01.001
[12] Shin H C, Orton M R, Collins D J, et al. Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2013, 35(8): 1930-1943. doi: 10.1109/TPAMI.2012.277
[13] Wang Z, Yang J. Automated detection of diabetic retinopathy using deep convolutional neural networks[J]. Medical Physics, 2016, 43(6): 3406. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=10.1118/1.4955912
[14] Kooi T, Litjens G, Van Ginneken B, et al. Large scale deep learning for computer aided detection of mammographic lesions[J]. Medical Image Analysis, 2017, 35: 303-312. doi: 10.1016/j.media.2016.07.007
[15] 陶永鹏, 景雨, 顼聪.融合超像素和CNN的CT图像分割方法[J].计算机工程与应用, 2019: 1-8.
Tao Y P, Jing Y, Xu C. CT image segmentation method combining superpixel and CNN[J]. Computer Engineering and Applications, 2019: 1-8.
[16] Yu L Q, Yang X, Hao C, et al. Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images[C]//Proceedings of the 31th AAAI Conference on Artificial Intelligence (AAAI-17), 2017: 66-72.
[17] Sevastopolsky A. Optic disc and cup segmentation methods for glaucoma detection with modification of U-Net convolutional neural network[J]. Pattern Recognition and Image Analysis, 2017, 27(3): 618-624. doi: 10.1134/S1054661817030269
[18] Milletari F, Navab N, Ahmadi S A. V-Net: fully convolutional neural networks for volumetric medical image segmentation[C]//2016 Fourth International Conference on 3D Vision (3DV), 2016: 565-571.
[19] Ren M. Learning a classification model for segmentation[C]//Proceedings Ninth IEEE International Conference on Computer Vision, 2003: 10-17.
[20] Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition[Z]. arXiv: 1409.1556, 2015.
[21] Aghaei F, Ross S R, Wang Y Z, et al. Implementation of a computer-aided detection tool for quantification of intracranial radiologic markers on brain CT images[C]//Proceedings Volume 10138, Medical Imaging 2017: Imaging Informatics for Healthcare, Research, and Applications, 2017: 10138.
[22] Korez R, Ibragimov B, Likar B, et al. Interpolation-based shape-constrained deformable model approach for segmentation of vertebrae from CT spine images[C]//Recent Advances in Computational Methods and Clinical Applications for Spine Imaging, 2015: 235-240.
[23] Liu X M, Guo S X, Yang B T, et al. Automatic organ segmentation for CT scans based on super-pixel and convolutional neural networks[J]. Journal of Digital Imaging, 2018, 31(5): 748-760. doi: 10.1007/s10278-018-0052-4
-
访问统计

 E-mail Alert
E-mail Alert RSS
RSS
 下载:
下载: